User login
Why like attracts like in malaria

Photo courtesy of CDC
Past research has shown that malaria-carrying mosquitoes prefer to feed on humans who are already infected with malaria.
Now, researchers believe they have discovered why.
The
team identified a naturally occurring compound, known as HMBPP, that is derived from the malaria parasite Plasmodium falciparum and
triggers the release of mosquito-attracting chemicals, making a human
host more enticing to the insects.
“The malaria parasite produces a molecule, HMBPP, which stimulates the human red blood cells to release more carbon dioxide and volatile compounds with an irresistible smell to malaria mosquitoes,” explained study author Ingrid Faye, of Stockholm University in Sweden.
She and her colleagues described this discovery in Science.
To determine if HMBPP could influence mosquitoes’ blood-meal-seeking and feeding behaviors, the researchers devised a “dual choice attraction” test for Anopheles gambiae mosquitoes.
Specifically, the team evaluated the mosquitoes’ preference to land on and feed off of an artificial membrane containing HMBPP-supplemented red blood cells (hmbRBCs) or normal red blood cells (RBCs).
More than 95% of the mosquitoes tested chose hmbRBCs over RBCs, and the mosquitoes consumed hmbRBCs more intensively and for longer periods of time than they did RBCs.
The researchers also found that blood spiked with HMBPP activated the expression of Plasmodium-specific genes involved in protecting the mosquitoes’ vital functions while also improving receptivity to infection and amplifying the likelihood of parasite transmission.
“This seems to be a well-functioning system, developed over millions of years, which means that the malaria parasite can survive and spread to more people without killing the host,” Faye said.
She and her colleagues believe the discovery of HMBPP as a driver of mosquito attraction exposes a weakness in Plasmodium that could be exploited to better pinpoint disease-carrying Anopheles gambiae mosquitoes and possibly prevent the spread of malaria. ![]()

Photo courtesy of CDC
Past research has shown that malaria-carrying mosquitoes prefer to feed on humans who are already infected with malaria.
Now, researchers believe they have discovered why.
The
team identified a naturally occurring compound, known as HMBPP, that is derived from the malaria parasite Plasmodium falciparum and
triggers the release of mosquito-attracting chemicals, making a human
host more enticing to the insects.
“The malaria parasite produces a molecule, HMBPP, which stimulates the human red blood cells to release more carbon dioxide and volatile compounds with an irresistible smell to malaria mosquitoes,” explained study author Ingrid Faye, of Stockholm University in Sweden.
She and her colleagues described this discovery in Science.
To determine if HMBPP could influence mosquitoes’ blood-meal-seeking and feeding behaviors, the researchers devised a “dual choice attraction” test for Anopheles gambiae mosquitoes.
Specifically, the team evaluated the mosquitoes’ preference to land on and feed off of an artificial membrane containing HMBPP-supplemented red blood cells (hmbRBCs) or normal red blood cells (RBCs).
More than 95% of the mosquitoes tested chose hmbRBCs over RBCs, and the mosquitoes consumed hmbRBCs more intensively and for longer periods of time than they did RBCs.
The researchers also found that blood spiked with HMBPP activated the expression of Plasmodium-specific genes involved in protecting the mosquitoes’ vital functions while also improving receptivity to infection and amplifying the likelihood of parasite transmission.
“This seems to be a well-functioning system, developed over millions of years, which means that the malaria parasite can survive and spread to more people without killing the host,” Faye said.
She and her colleagues believe the discovery of HMBPP as a driver of mosquito attraction exposes a weakness in Plasmodium that could be exploited to better pinpoint disease-carrying Anopheles gambiae mosquitoes and possibly prevent the spread of malaria. ![]()

Photo courtesy of CDC
Past research has shown that malaria-carrying mosquitoes prefer to feed on humans who are already infected with malaria.
Now, researchers believe they have discovered why.
The
team identified a naturally occurring compound, known as HMBPP, that is derived from the malaria parasite Plasmodium falciparum and
triggers the release of mosquito-attracting chemicals, making a human
host more enticing to the insects.
“The malaria parasite produces a molecule, HMBPP, which stimulates the human red blood cells to release more carbon dioxide and volatile compounds with an irresistible smell to malaria mosquitoes,” explained study author Ingrid Faye, of Stockholm University in Sweden.
She and her colleagues described this discovery in Science.
To determine if HMBPP could influence mosquitoes’ blood-meal-seeking and feeding behaviors, the researchers devised a “dual choice attraction” test for Anopheles gambiae mosquitoes.
Specifically, the team evaluated the mosquitoes’ preference to land on and feed off of an artificial membrane containing HMBPP-supplemented red blood cells (hmbRBCs) or normal red blood cells (RBCs).
More than 95% of the mosquitoes tested chose hmbRBCs over RBCs, and the mosquitoes consumed hmbRBCs more intensively and for longer periods of time than they did RBCs.
The researchers also found that blood spiked with HMBPP activated the expression of Plasmodium-specific genes involved in protecting the mosquitoes’ vital functions while also improving receptivity to infection and amplifying the likelihood of parasite transmission.
“This seems to be a well-functioning system, developed over millions of years, which means that the malaria parasite can survive and spread to more people without killing the host,” Faye said.
She and her colleagues believe the discovery of HMBPP as a driver of mosquito attraction exposes a weakness in Plasmodium that could be exploited to better pinpoint disease-carrying Anopheles gambiae mosquitoes and possibly prevent the spread of malaria. ![]()
Computer predicts remission, relapse in AML

Photo by Darren Baker
Researchers say they have developed the first computer machine-learning model to accurately predict which patients diagnosed with acute
myeloid leukemia (AML) will go into remission after treatment and which will relapse.
“It’s pretty straightforward to teach a computer to recognize AML, once you develop a robust algorithm, and, in previous work, we did it with almost 100% accuracy,” said study author Murat Dundar, PhD of Indiana University-Purdue University Indianapolis.
“What was challenging was to go beyond that work and teach the computer to accurately predict the direction of change in disease progression in AML patients, interpreting new data to predict the unknown: which new AML patients will go into remission and which will relapse.”
Dr Dundar and his colleagues described this work in IEEE Transactions on Biomedical Engineering.
The researchers said they modeled data from multiple flow cytometry samples to identify functionally distinct cell populations and their local realizations. Each sample was characterized by the proportions of recovered cell populations, which were used to predict the direction of change in disease progression for each AML patient.
“As the input, our computational system employs data from flow cytometry, a widely utilized technology that can rapidly provide detailed characteristics of single cells in samples such as blood or bone marrow,” explained study author Bartek Rajwa, PhD, of Purdue University in Lafayette, Indiana.
“Traditionally, the results of flow cytometry analyses are evaluated by highly trained human experts rather than by machine-learning algorithms. But computers are often better at extracting knowledge from complex data than humans are.”
The researchers used 200 diseased and non-diseased immunophenotypic panels for training and tested
the computational system with samples collected at multiple time points from 36 additional AML patients.
The system was able to predict remission with 100% accuracy (26 of 26 cases) and relapse with 90% accuracy (9 of 10 cases).
“Machine learning is not about modeling data,” Dr Dundar noted. “It’s about extracting knowledge from the data you have so you can build a powerful, intuitive tool that can make predictions about future data that the computer has not previously seen. The machine is learning, not memorizing, and that’s what we did.” ![]()

Photo by Darren Baker
Researchers say they have developed the first computer machine-learning model to accurately predict which patients diagnosed with acute
myeloid leukemia (AML) will go into remission after treatment and which will relapse.
“It’s pretty straightforward to teach a computer to recognize AML, once you develop a robust algorithm, and, in previous work, we did it with almost 100% accuracy,” said study author Murat Dundar, PhD of Indiana University-Purdue University Indianapolis.
“What was challenging was to go beyond that work and teach the computer to accurately predict the direction of change in disease progression in AML patients, interpreting new data to predict the unknown: which new AML patients will go into remission and which will relapse.”
Dr Dundar and his colleagues described this work in IEEE Transactions on Biomedical Engineering.
The researchers said they modeled data from multiple flow cytometry samples to identify functionally distinct cell populations and their local realizations. Each sample was characterized by the proportions of recovered cell populations, which were used to predict the direction of change in disease progression for each AML patient.
“As the input, our computational system employs data from flow cytometry, a widely utilized technology that can rapidly provide detailed characteristics of single cells in samples such as blood or bone marrow,” explained study author Bartek Rajwa, PhD, of Purdue University in Lafayette, Indiana.
“Traditionally, the results of flow cytometry analyses are evaluated by highly trained human experts rather than by machine-learning algorithms. But computers are often better at extracting knowledge from complex data than humans are.”
The researchers used 200 diseased and non-diseased immunophenotypic panels for training and tested
the computational system with samples collected at multiple time points from 36 additional AML patients.
The system was able to predict remission with 100% accuracy (26 of 26 cases) and relapse with 90% accuracy (9 of 10 cases).
“Machine learning is not about modeling data,” Dr Dundar noted. “It’s about extracting knowledge from the data you have so you can build a powerful, intuitive tool that can make predictions about future data that the computer has not previously seen. The machine is learning, not memorizing, and that’s what we did.” ![]()

Photo by Darren Baker
Researchers say they have developed the first computer machine-learning model to accurately predict which patients diagnosed with acute
myeloid leukemia (AML) will go into remission after treatment and which will relapse.
“It’s pretty straightforward to teach a computer to recognize AML, once you develop a robust algorithm, and, in previous work, we did it with almost 100% accuracy,” said study author Murat Dundar, PhD of Indiana University-Purdue University Indianapolis.
“What was challenging was to go beyond that work and teach the computer to accurately predict the direction of change in disease progression in AML patients, interpreting new data to predict the unknown: which new AML patients will go into remission and which will relapse.”
Dr Dundar and his colleagues described this work in IEEE Transactions on Biomedical Engineering.
The researchers said they modeled data from multiple flow cytometry samples to identify functionally distinct cell populations and their local realizations. Each sample was characterized by the proportions of recovered cell populations, which were used to predict the direction of change in disease progression for each AML patient.
“As the input, our computational system employs data from flow cytometry, a widely utilized technology that can rapidly provide detailed characteristics of single cells in samples such as blood or bone marrow,” explained study author Bartek Rajwa, PhD, of Purdue University in Lafayette, Indiana.
“Traditionally, the results of flow cytometry analyses are evaluated by highly trained human experts rather than by machine-learning algorithms. But computers are often better at extracting knowledge from complex data than humans are.”
The researchers used 200 diseased and non-diseased immunophenotypic panels for training and tested
the computational system with samples collected at multiple time points from 36 additional AML patients.
The system was able to predict remission with 100% accuracy (26 of 26 cases) and relapse with 90% accuracy (9 of 10 cases).
“Machine learning is not about modeling data,” Dr Dundar noted. “It’s about extracting knowledge from the data you have so you can build a powerful, intuitive tool that can make predictions about future data that the computer has not previously seen. The machine is learning, not memorizing, and that’s what we did.” ![]()
Genetic profiling can guide HSCT in MDS, team says
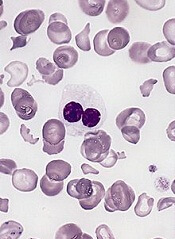
Genetic profiling can be used to determine which patients with myelodysplastic syndrome (MDS) are likely to benefit from allogeneic hematopoietic stem cell transplant (HSCT), according to research published in NEJM.
Targeted sequencing of 129 genes revealed mutations that, after adjustment for clinical variables, were associated with shorter survival and/or relapse after HSCT.
Patients with mutations in TP53, JAK2, and the RAS pathway tended to have worse outcomes after HSCT than patients without such mutations.
“Although donor stem cell transplantation is the only curative therapy for MDS, many patients die after transplantation, largely due to relapse of the disease or complications relating to the transplant itself,” said study author R. Coleman Lindsley, MD, PhD, of Dana-Farber Cancer Institute in Boston, Massachusetts.
“As physicians, one of our major challenges is to be able to predict which patients are most likely to benefit from a transplant. Improving our ability to identify patients who are most likely to have a relapse or to experience life-threatening complications from a transplant could lead to better pre-transplant therapies and strategies for preventing relapse.”
Researchers have long known that specific genetic mutations are closely related to the course MDS takes. With this study, Dr Lindsley and his colleagues sought to discover whether mutations can be used to predict how patients will fare following allogeneic HSCT.
The team analyzed blood samples from 1514 MDS patients, performing targeted sequencing of 129 genes. The genes were selected based on their known or suspected involvement in the pathogenesis of myeloid cancers or bone marrow failure syndromes.
Dr Lindsley and his colleagues then evaluated the association between mutations and HSCT outcomes, including overall survival, relapse, and death without relapse.
After adjusting for significant clinical variables, the researchers found that having mutated TP53 was significantly associated with shorter survival and shorter time to relapse after HSCT (P<0.001 for both comparisons). This was true whether patients received standard conditioning or reduced-intensity conditioning.
In patients age 40 and older who did not have TP53 mutations, mutations in RAS pathway genes (P=0.004) or JAK2 (P=0.001) were significantly associated with shorter survival.
The shorter survival in patients with mutated RAS pathway genes was due to a higher risk of relapse, while the shorter survival in patients with JAK2 mutations was due to a higher risk of death without relapse.
In contrast to TP53 mutations, the adverse effect of RAS mutations on survival and risk of relapse was evident only in patients who received reduced-intensity conditioning (P<0.001). This suggests these patients may benefit from higher intensity conditioning regimens, the researchers said.
This study also yielded insights about the biology of MDS in specific groups of patients.
For example, the researchers found that 4% of MDS patients between the ages of 18 and 40 had mutations associated with Shwachman-Diamond syndrome (in the SBDS gene), but most of them had not previously been diagnosed with the syndrome.
In each case, the patients had acquired a TP53 mutation, suggesting not only how MDS develops in patients with Schwachman-Diamond syndrome but also what underlies their poor prognosis after HSCT.
The researchers also analyzed patients with therapy-related MDS. The team found that TP53 mutations and mutations in PPM1D, a gene that regulates TP53 function, were far more common in these patients than in those with primary MDS (15% and 3%, respectively, P<0.001).
“In deciding whether a stem cell transplant is appropriate for a patient with MDS, it’s always necessary to balance the potential benefit with the risk of complications,” Dr Lindsley noted.
“Our findings offer physicians a guide—based on the genetic profile of the disease and certain clinical factors—to identifying patients for whom a transplant is appropriate, and the intensity of treatment most likely to be effective.” ![]()

Genetic profiling can be used to determine which patients with myelodysplastic syndrome (MDS) are likely to benefit from allogeneic hematopoietic stem cell transplant (HSCT), according to research published in NEJM.
Targeted sequencing of 129 genes revealed mutations that, after adjustment for clinical variables, were associated with shorter survival and/or relapse after HSCT.
Patients with mutations in TP53, JAK2, and the RAS pathway tended to have worse outcomes after HSCT than patients without such mutations.
“Although donor stem cell transplantation is the only curative therapy for MDS, many patients die after transplantation, largely due to relapse of the disease or complications relating to the transplant itself,” said study author R. Coleman Lindsley, MD, PhD, of Dana-Farber Cancer Institute in Boston, Massachusetts.
“As physicians, one of our major challenges is to be able to predict which patients are most likely to benefit from a transplant. Improving our ability to identify patients who are most likely to have a relapse or to experience life-threatening complications from a transplant could lead to better pre-transplant therapies and strategies for preventing relapse.”
Researchers have long known that specific genetic mutations are closely related to the course MDS takes. With this study, Dr Lindsley and his colleagues sought to discover whether mutations can be used to predict how patients will fare following allogeneic HSCT.
The team analyzed blood samples from 1514 MDS patients, performing targeted sequencing of 129 genes. The genes were selected based on their known or suspected involvement in the pathogenesis of myeloid cancers or bone marrow failure syndromes.
Dr Lindsley and his colleagues then evaluated the association between mutations and HSCT outcomes, including overall survival, relapse, and death without relapse.
After adjusting for significant clinical variables, the researchers found that having mutated TP53 was significantly associated with shorter survival and shorter time to relapse after HSCT (P<0.001 for both comparisons). This was true whether patients received standard conditioning or reduced-intensity conditioning.
In patients age 40 and older who did not have TP53 mutations, mutations in RAS pathway genes (P=0.004) or JAK2 (P=0.001) were significantly associated with shorter survival.
The shorter survival in patients with mutated RAS pathway genes was due to a higher risk of relapse, while the shorter survival in patients with JAK2 mutations was due to a higher risk of death without relapse.
In contrast to TP53 mutations, the adverse effect of RAS mutations on survival and risk of relapse was evident only in patients who received reduced-intensity conditioning (P<0.001). This suggests these patients may benefit from higher intensity conditioning regimens, the researchers said.
This study also yielded insights about the biology of MDS in specific groups of patients.
For example, the researchers found that 4% of MDS patients between the ages of 18 and 40 had mutations associated with Shwachman-Diamond syndrome (in the SBDS gene), but most of them had not previously been diagnosed with the syndrome.
In each case, the patients had acquired a TP53 mutation, suggesting not only how MDS develops in patients with Schwachman-Diamond syndrome but also what underlies their poor prognosis after HSCT.
The researchers also analyzed patients with therapy-related MDS. The team found that TP53 mutations and mutations in PPM1D, a gene that regulates TP53 function, were far more common in these patients than in those with primary MDS (15% and 3%, respectively, P<0.001).
“In deciding whether a stem cell transplant is appropriate for a patient with MDS, it’s always necessary to balance the potential benefit with the risk of complications,” Dr Lindsley noted.
“Our findings offer physicians a guide—based on the genetic profile of the disease and certain clinical factors—to identifying patients for whom a transplant is appropriate, and the intensity of treatment most likely to be effective.” ![]()

Genetic profiling can be used to determine which patients with myelodysplastic syndrome (MDS) are likely to benefit from allogeneic hematopoietic stem cell transplant (HSCT), according to research published in NEJM.
Targeted sequencing of 129 genes revealed mutations that, after adjustment for clinical variables, were associated with shorter survival and/or relapse after HSCT.
Patients with mutations in TP53, JAK2, and the RAS pathway tended to have worse outcomes after HSCT than patients without such mutations.
“Although donor stem cell transplantation is the only curative therapy for MDS, many patients die after transplantation, largely due to relapse of the disease or complications relating to the transplant itself,” said study author R. Coleman Lindsley, MD, PhD, of Dana-Farber Cancer Institute in Boston, Massachusetts.
“As physicians, one of our major challenges is to be able to predict which patients are most likely to benefit from a transplant. Improving our ability to identify patients who are most likely to have a relapse or to experience life-threatening complications from a transplant could lead to better pre-transplant therapies and strategies for preventing relapse.”
Researchers have long known that specific genetic mutations are closely related to the course MDS takes. With this study, Dr Lindsley and his colleagues sought to discover whether mutations can be used to predict how patients will fare following allogeneic HSCT.
The team analyzed blood samples from 1514 MDS patients, performing targeted sequencing of 129 genes. The genes were selected based on their known or suspected involvement in the pathogenesis of myeloid cancers or bone marrow failure syndromes.
Dr Lindsley and his colleagues then evaluated the association between mutations and HSCT outcomes, including overall survival, relapse, and death without relapse.
After adjusting for significant clinical variables, the researchers found that having mutated TP53 was significantly associated with shorter survival and shorter time to relapse after HSCT (P<0.001 for both comparisons). This was true whether patients received standard conditioning or reduced-intensity conditioning.
In patients age 40 and older who did not have TP53 mutations, mutations in RAS pathway genes (P=0.004) or JAK2 (P=0.001) were significantly associated with shorter survival.
The shorter survival in patients with mutated RAS pathway genes was due to a higher risk of relapse, while the shorter survival in patients with JAK2 mutations was due to a higher risk of death without relapse.
In contrast to TP53 mutations, the adverse effect of RAS mutations on survival and risk of relapse was evident only in patients who received reduced-intensity conditioning (P<0.001). This suggests these patients may benefit from higher intensity conditioning regimens, the researchers said.
This study also yielded insights about the biology of MDS in specific groups of patients.
For example, the researchers found that 4% of MDS patients between the ages of 18 and 40 had mutations associated with Shwachman-Diamond syndrome (in the SBDS gene), but most of them had not previously been diagnosed with the syndrome.
In each case, the patients had acquired a TP53 mutation, suggesting not only how MDS develops in patients with Schwachman-Diamond syndrome but also what underlies their poor prognosis after HSCT.
The researchers also analyzed patients with therapy-related MDS. The team found that TP53 mutations and mutations in PPM1D, a gene that regulates TP53 function, were far more common in these patients than in those with primary MDS (15% and 3%, respectively, P<0.001).
“In deciding whether a stem cell transplant is appropriate for a patient with MDS, it’s always necessary to balance the potential benefit with the risk of complications,” Dr Lindsley noted.
“Our findings offer physicians a guide—based on the genetic profile of the disease and certain clinical factors—to identifying patients for whom a transplant is appropriate, and the intensity of treatment most likely to be effective.” ![]()
Algorithm predicts NRM, GVHD after HSCT
A biomarker algorithm can identify patients with a high risk of graft-vs-host disease (GVHD) and non-relapse mortality (NRM) after hematopoietic stem cell transplant (HSCT), according to researchers.
The team found evidence to suggest that 2 proteins—ST2 and REG3α—present in blood drawn a week after HSCT can predict the likelihood of GVHD, including lethal GVHD, and NRM in patients with hematologic disorders.
James L.M. Ferrara, MD, of the Icahn School of Medicine at Mount Sinai in New York, New York, and his colleagues reported these findings in JCI Insight.
The researchers analyzed blood samples collected on day 7 after HSCT from 1287 patients. Of these, 620 samples were designated the training set.
The team measured the concentrations of 4 GVHD biomarkers—ST2, REG3α, TNFR1, and IL-2Rα—in the training set and used them to model 6-month NRM in an attempt to identify the best algorithm that defined 2 distinct risk groups.
The researchers applied the resulting algorithm to the test set of samples (n=309) and the validation set of samples (n=358).
The final algorithm used ST2 and REG3α concentrations to identify patients with a high and low risk of NRM at 6 months. Sixteen percent of patients in the training set belonged to the high-risk group, as did 17% of the test set and 20% of the validation set.
In the training set, the cumulative incidence of NRM at 6 months was 28% in the high-risk group and 7% in the low-risk group (P<0.001). The incidence was 33% and 7%, respectively (P<0.001), in the test set and 26% and 10%, respectively (P<0.001), in the validation set.
The high-risk patients were 3 times more likely to die from GVHD than low-risk patients in the overall cohort. The incidence of lethal GVHD was 19% and 6%, respectively (P<0.001).
GVHD-related mortality in the high-risk and low-risk groups, respectively, was 18% and 5% (P<0.001) in the training set, 24% and 4% (P<0.001) in the test set, and 14% and 5% (P<0.001) in the validation set.
The researchers said their algorithm can also be adapted to define 3 distinct risk groups at GVHD onset—Ann Arbor scores 1, 2, and 3.
The team dubbed their algorithm the “MAGIC algorithm,” after the Mount Sinai Acute GVHD International Consortium (MAGIC).
“The MAGIC algorithm gives doctors a roadmap to save many lives in the future,” Dr Ferrara said. “This simple blood test can determine which bone marrow transplant patients are at high risk for a lethal complication before it occurs. It will allow early intervention and potentially save many lives.”
Doctors at Mount Sinai are now designing clinical trials to determine whether immunotherapy drugs would benefit patients if the MAGIC algorithm determines they are at high risk for severe GVHD.
The researchers believe that if patients receive the drugs once the blood test is administered, which is well before symptoms develop, they would be spared the full force of GVHD, and fewer of them would die.
“This test will make bone marrow transplant safer and more effective for patients because it will guide adjustment of medications to protect against graft-vs-host disease,” said study author John Levine, MD, of the Icahn School of Medicine at Mount Sinai.
“If successful, the early use of the drugs would become a standard of care for bone marrow transplant patients.” ![]()
A biomarker algorithm can identify patients with a high risk of graft-vs-host disease (GVHD) and non-relapse mortality (NRM) after hematopoietic stem cell transplant (HSCT), according to researchers.
The team found evidence to suggest that 2 proteins—ST2 and REG3α—present in blood drawn a week after HSCT can predict the likelihood of GVHD, including lethal GVHD, and NRM in patients with hematologic disorders.
James L.M. Ferrara, MD, of the Icahn School of Medicine at Mount Sinai in New York, New York, and his colleagues reported these findings in JCI Insight.
The researchers analyzed blood samples collected on day 7 after HSCT from 1287 patients. Of these, 620 samples were designated the training set.
The team measured the concentrations of 4 GVHD biomarkers—ST2, REG3α, TNFR1, and IL-2Rα—in the training set and used them to model 6-month NRM in an attempt to identify the best algorithm that defined 2 distinct risk groups.
The researchers applied the resulting algorithm to the test set of samples (n=309) and the validation set of samples (n=358).
The final algorithm used ST2 and REG3α concentrations to identify patients with a high and low risk of NRM at 6 months. Sixteen percent of patients in the training set belonged to the high-risk group, as did 17% of the test set and 20% of the validation set.
In the training set, the cumulative incidence of NRM at 6 months was 28% in the high-risk group and 7% in the low-risk group (P<0.001). The incidence was 33% and 7%, respectively (P<0.001), in the test set and 26% and 10%, respectively (P<0.001), in the validation set.
The high-risk patients were 3 times more likely to die from GVHD than low-risk patients in the overall cohort. The incidence of lethal GVHD was 19% and 6%, respectively (P<0.001).
GVHD-related mortality in the high-risk and low-risk groups, respectively, was 18% and 5% (P<0.001) in the training set, 24% and 4% (P<0.001) in the test set, and 14% and 5% (P<0.001) in the validation set.
The researchers said their algorithm can also be adapted to define 3 distinct risk groups at GVHD onset—Ann Arbor scores 1, 2, and 3.
The team dubbed their algorithm the “MAGIC algorithm,” after the Mount Sinai Acute GVHD International Consortium (MAGIC).
“The MAGIC algorithm gives doctors a roadmap to save many lives in the future,” Dr Ferrara said. “This simple blood test can determine which bone marrow transplant patients are at high risk for a lethal complication before it occurs. It will allow early intervention and potentially save many lives.”
Doctors at Mount Sinai are now designing clinical trials to determine whether immunotherapy drugs would benefit patients if the MAGIC algorithm determines they are at high risk for severe GVHD.
The researchers believe that if patients receive the drugs once the blood test is administered, which is well before symptoms develop, they would be spared the full force of GVHD, and fewer of them would die.
“This test will make bone marrow transplant safer and more effective for patients because it will guide adjustment of medications to protect against graft-vs-host disease,” said study author John Levine, MD, of the Icahn School of Medicine at Mount Sinai.
“If successful, the early use of the drugs would become a standard of care for bone marrow transplant patients.” ![]()
A biomarker algorithm can identify patients with a high risk of graft-vs-host disease (GVHD) and non-relapse mortality (NRM) after hematopoietic stem cell transplant (HSCT), according to researchers.
The team found evidence to suggest that 2 proteins—ST2 and REG3α—present in blood drawn a week after HSCT can predict the likelihood of GVHD, including lethal GVHD, and NRM in patients with hematologic disorders.
James L.M. Ferrara, MD, of the Icahn School of Medicine at Mount Sinai in New York, New York, and his colleagues reported these findings in JCI Insight.
The researchers analyzed blood samples collected on day 7 after HSCT from 1287 patients. Of these, 620 samples were designated the training set.
The team measured the concentrations of 4 GVHD biomarkers—ST2, REG3α, TNFR1, and IL-2Rα—in the training set and used them to model 6-month NRM in an attempt to identify the best algorithm that defined 2 distinct risk groups.
The researchers applied the resulting algorithm to the test set of samples (n=309) and the validation set of samples (n=358).
The final algorithm used ST2 and REG3α concentrations to identify patients with a high and low risk of NRM at 6 months. Sixteen percent of patients in the training set belonged to the high-risk group, as did 17% of the test set and 20% of the validation set.
In the training set, the cumulative incidence of NRM at 6 months was 28% in the high-risk group and 7% in the low-risk group (P<0.001). The incidence was 33% and 7%, respectively (P<0.001), in the test set and 26% and 10%, respectively (P<0.001), in the validation set.
The high-risk patients were 3 times more likely to die from GVHD than low-risk patients in the overall cohort. The incidence of lethal GVHD was 19% and 6%, respectively (P<0.001).
GVHD-related mortality in the high-risk and low-risk groups, respectively, was 18% and 5% (P<0.001) in the training set, 24% and 4% (P<0.001) in the test set, and 14% and 5% (P<0.001) in the validation set.
The researchers said their algorithm can also be adapted to define 3 distinct risk groups at GVHD onset—Ann Arbor scores 1, 2, and 3.
The team dubbed their algorithm the “MAGIC algorithm,” after the Mount Sinai Acute GVHD International Consortium (MAGIC).
“The MAGIC algorithm gives doctors a roadmap to save many lives in the future,” Dr Ferrara said. “This simple blood test can determine which bone marrow transplant patients are at high risk for a lethal complication before it occurs. It will allow early intervention and potentially save many lives.”
Doctors at Mount Sinai are now designing clinical trials to determine whether immunotherapy drugs would benefit patients if the MAGIC algorithm determines they are at high risk for severe GVHD.
The researchers believe that if patients receive the drugs once the blood test is administered, which is well before symptoms develop, they would be spared the full force of GVHD, and fewer of them would die.
“This test will make bone marrow transplant safer and more effective for patients because it will guide adjustment of medications to protect against graft-vs-host disease,” said study author John Levine, MD, of the Icahn School of Medicine at Mount Sinai.
“If successful, the early use of the drugs would become a standard of care for bone marrow transplant patients.” ![]()
Study shows no increased risk of mutations with iPSCs

Image from Salk Institute
The use of induced pluripotent stem cells (iPSCs) in biomedical research and medicine has been slowed by concerns that these cells are prone to increased numbers of genetic mutations.
However, a new study suggests iPSCs do not develop more mutations than cells that are duplicated by subcloning, a technique where single cells are cultured individually and then grown into a cell line.
Subcloning is similar to the technique used to create iPSCs, except the subcloned cells are not treated with the reprogramming factors that have been thought to cause mutations in iPSCs.
“These findings suggest that the question of safety shouldn’t impede research using iPSCs,” said study author Paul Liu, MD, PhD, of the National Human Genome Research Institute, part of the National Institutes of Health, in Bethesda, Maryland.
Dr Liu and his colleagues reported the findings in PNAS.
For this study, the researchers examined 2 sets of donated cells. One set came from a healthy individual, and the second came from a person with familial platelet disorder.
Using fibroblasts from each of the donors, the researchers created genetically identical copies of the cells using both the iPSC and subcloning techniques.
The team then sequenced the DNA of the fibroblasts as well as the iPSCs and the subcloned cells and determined that mutations occurred at the same rate in cells that were reprogrammed and cells that were subcloned.
More than 90% of the genetic variants detected in the iPSCs and subclones were rare variants inherited from the parent cells.
This suggests that most mutations in iPSCs are not generated during the reprogramming or iPSC production phase and provides evidence that iPSCs are stable and safe to use for both basic and clinical research, Dr Liu said.
“Based on this data, we plan to start using iPSCs to gain a deeper understanding of how diseases start and progress,” said study author Erika Mijin Kwon, PhD, also of the National Human Genome Research Institute.
“We eventually hope to develop new therapies to treat patients with leukemia using their own iPSCs. We encourage other researchers to embrace the use of iPSCs.” ![]()

Image from Salk Institute
The use of induced pluripotent stem cells (iPSCs) in biomedical research and medicine has been slowed by concerns that these cells are prone to increased numbers of genetic mutations.
However, a new study suggests iPSCs do not develop more mutations than cells that are duplicated by subcloning, a technique where single cells are cultured individually and then grown into a cell line.
Subcloning is similar to the technique used to create iPSCs, except the subcloned cells are not treated with the reprogramming factors that have been thought to cause mutations in iPSCs.
“These findings suggest that the question of safety shouldn’t impede research using iPSCs,” said study author Paul Liu, MD, PhD, of the National Human Genome Research Institute, part of the National Institutes of Health, in Bethesda, Maryland.
Dr Liu and his colleagues reported the findings in PNAS.
For this study, the researchers examined 2 sets of donated cells. One set came from a healthy individual, and the second came from a person with familial platelet disorder.
Using fibroblasts from each of the donors, the researchers created genetically identical copies of the cells using both the iPSC and subcloning techniques.
The team then sequenced the DNA of the fibroblasts as well as the iPSCs and the subcloned cells and determined that mutations occurred at the same rate in cells that were reprogrammed and cells that were subcloned.
More than 90% of the genetic variants detected in the iPSCs and subclones were rare variants inherited from the parent cells.
This suggests that most mutations in iPSCs are not generated during the reprogramming or iPSC production phase and provides evidence that iPSCs are stable and safe to use for both basic and clinical research, Dr Liu said.
“Based on this data, we plan to start using iPSCs to gain a deeper understanding of how diseases start and progress,” said study author Erika Mijin Kwon, PhD, also of the National Human Genome Research Institute.
“We eventually hope to develop new therapies to treat patients with leukemia using their own iPSCs. We encourage other researchers to embrace the use of iPSCs.” ![]()

Image from Salk Institute
The use of induced pluripotent stem cells (iPSCs) in biomedical research and medicine has been slowed by concerns that these cells are prone to increased numbers of genetic mutations.
However, a new study suggests iPSCs do not develop more mutations than cells that are duplicated by subcloning, a technique where single cells are cultured individually and then grown into a cell line.
Subcloning is similar to the technique used to create iPSCs, except the subcloned cells are not treated with the reprogramming factors that have been thought to cause mutations in iPSCs.
“These findings suggest that the question of safety shouldn’t impede research using iPSCs,” said study author Paul Liu, MD, PhD, of the National Human Genome Research Institute, part of the National Institutes of Health, in Bethesda, Maryland.
Dr Liu and his colleagues reported the findings in PNAS.
For this study, the researchers examined 2 sets of donated cells. One set came from a healthy individual, and the second came from a person with familial platelet disorder.
Using fibroblasts from each of the donors, the researchers created genetically identical copies of the cells using both the iPSC and subcloning techniques.
The team then sequenced the DNA of the fibroblasts as well as the iPSCs and the subcloned cells and determined that mutations occurred at the same rate in cells that were reprogrammed and cells that were subcloned.
More than 90% of the genetic variants detected in the iPSCs and subclones were rare variants inherited from the parent cells.
This suggests that most mutations in iPSCs are not generated during the reprogramming or iPSC production phase and provides evidence that iPSCs are stable and safe to use for both basic and clinical research, Dr Liu said.
“Based on this data, we plan to start using iPSCs to gain a deeper understanding of how diseases start and progress,” said study author Erika Mijin Kwon, PhD, also of the National Human Genome Research Institute.
“We eventually hope to develop new therapies to treat patients with leukemia using their own iPSCs. We encourage other researchers to embrace the use of iPSCs.” ![]()
iPSCs used to identify potential treatment for DBA
from cells from a DBA patient
Image courtesy of
Boston Children’s Hospital
Researchers have used induced pluripotent stem cells (iPSCs) to identify a compound that could treat Diamond Blackfan anemia (DBA).
The team used iPSCs to generate expandable hematopoietic progenitor cells (HPCs) that recapitulate the defects in erythroid differentiation observed in patients with DBA.
The researchers then used the HPCs to screen chemical compounds that might be used to treat DBA.
One of these compounds, SMER28, enhanced erythropoiesis in models of DBA.
“It is very satisfying as physician-scientists to find new potential treatments for rare blood diseases such as Diamond Blackfan anemia,” said study author Leonard Zon, MD, of Boston Children’s Hospital and Dana-Farber Cancer Institute in Boston, Massachusetts.
Dr Zon and his colleagues described this work in Science Translational Medicine.
The researchers first took fibroblasts from 2 patients with DBA and reprogrammed them into iPSCs. From the iPSCs, the team generated HPCs, which they loaded into a high-throughput drug screening system.
Testing a library of 1440 chemicals, the researchers found several that showed promise for treating DBA in vitro. One compound, SMER28, was able to induce red blood cell (RBC) production in mice and zebrafish.
The researchers believe this study marks an important advance in the stem cell field.
“[iPSCs] have been hard to instruct when it comes to making blood,” said study author Sergei Doulatov, PhD, of the University of Washington in Seattle.
“This is the first time [iPSCs] have been used to identify a drug to treat a blood disorder.”
Making RBCs
As in DBA itself, the patient-derived HPCs, studied in vitro, failed to generate erythroid cells. The same was true when the cells were transplanted into mice.
However, the chemical screen got several “hits.” In wells loaded with certain chemicals, erythroid cells began appearing. Because of its especially strong effect, SMER28 was put through additional testing.
When SMER28 was used to treat the marrow in zebrafish and mouse models of DBA, the animals made erythroid progenitor cells that, in turn, made RBCs, reversing or stabilizing anemia. The same was true in cells from DBA patients transplanted into mice.
The higher the dose of SMER28, the more RBCs were produced, and no ill effects were found. However, formal toxicity studies have not been conducted.
SMER28 has been tested preclinically for some neurodegenerative diseases. It activates an autophagy pathway that recycles damaged cellular components.
In DBA, SMER28 appears to turn on autophagy in erythroid progenitors. Dr Doulatov plans to further explore how this interferes with RBC production. ![]()

from cells from a DBA patient
Image courtesy of
Boston Children’s Hospital
Researchers have used induced pluripotent stem cells (iPSCs) to identify a compound that could treat Diamond Blackfan anemia (DBA).
The team used iPSCs to generate expandable hematopoietic progenitor cells (HPCs) that recapitulate the defects in erythroid differentiation observed in patients with DBA.
The researchers then used the HPCs to screen chemical compounds that might be used to treat DBA.
One of these compounds, SMER28, enhanced erythropoiesis in models of DBA.
“It is very satisfying as physician-scientists to find new potential treatments for rare blood diseases such as Diamond Blackfan anemia,” said study author Leonard Zon, MD, of Boston Children’s Hospital and Dana-Farber Cancer Institute in Boston, Massachusetts.
Dr Zon and his colleagues described this work in Science Translational Medicine.
The researchers first took fibroblasts from 2 patients with DBA and reprogrammed them into iPSCs. From the iPSCs, the team generated HPCs, which they loaded into a high-throughput drug screening system.
Testing a library of 1440 chemicals, the researchers found several that showed promise for treating DBA in vitro. One compound, SMER28, was able to induce red blood cell (RBC) production in mice and zebrafish.
The researchers believe this study marks an important advance in the stem cell field.
“[iPSCs] have been hard to instruct when it comes to making blood,” said study author Sergei Doulatov, PhD, of the University of Washington in Seattle.
“This is the first time [iPSCs] have been used to identify a drug to treat a blood disorder.”
Making RBCs
As in DBA itself, the patient-derived HPCs, studied in vitro, failed to generate erythroid cells. The same was true when the cells were transplanted into mice.
However, the chemical screen got several “hits.” In wells loaded with certain chemicals, erythroid cells began appearing. Because of its especially strong effect, SMER28 was put through additional testing.
When SMER28 was used to treat the marrow in zebrafish and mouse models of DBA, the animals made erythroid progenitor cells that, in turn, made RBCs, reversing or stabilizing anemia. The same was true in cells from DBA patients transplanted into mice.
The higher the dose of SMER28, the more RBCs were produced, and no ill effects were found. However, formal toxicity studies have not been conducted.
SMER28 has been tested preclinically for some neurodegenerative diseases. It activates an autophagy pathway that recycles damaged cellular components.
In DBA, SMER28 appears to turn on autophagy in erythroid progenitors. Dr Doulatov plans to further explore how this interferes with RBC production. ![]()

from cells from a DBA patient
Image courtesy of
Boston Children’s Hospital
Researchers have used induced pluripotent stem cells (iPSCs) to identify a compound that could treat Diamond Blackfan anemia (DBA).
The team used iPSCs to generate expandable hematopoietic progenitor cells (HPCs) that recapitulate the defects in erythroid differentiation observed in patients with DBA.
The researchers then used the HPCs to screen chemical compounds that might be used to treat DBA.
One of these compounds, SMER28, enhanced erythropoiesis in models of DBA.
“It is very satisfying as physician-scientists to find new potential treatments for rare blood diseases such as Diamond Blackfan anemia,” said study author Leonard Zon, MD, of Boston Children’s Hospital and Dana-Farber Cancer Institute in Boston, Massachusetts.
Dr Zon and his colleagues described this work in Science Translational Medicine.
The researchers first took fibroblasts from 2 patients with DBA and reprogrammed them into iPSCs. From the iPSCs, the team generated HPCs, which they loaded into a high-throughput drug screening system.
Testing a library of 1440 chemicals, the researchers found several that showed promise for treating DBA in vitro. One compound, SMER28, was able to induce red blood cell (RBC) production in mice and zebrafish.
The researchers believe this study marks an important advance in the stem cell field.
“[iPSCs] have been hard to instruct when it comes to making blood,” said study author Sergei Doulatov, PhD, of the University of Washington in Seattle.
“This is the first time [iPSCs] have been used to identify a drug to treat a blood disorder.”
Making RBCs
As in DBA itself, the patient-derived HPCs, studied in vitro, failed to generate erythroid cells. The same was true when the cells were transplanted into mice.
However, the chemical screen got several “hits.” In wells loaded with certain chemicals, erythroid cells began appearing. Because of its especially strong effect, SMER28 was put through additional testing.
When SMER28 was used to treat the marrow in zebrafish and mouse models of DBA, the animals made erythroid progenitor cells that, in turn, made RBCs, reversing or stabilizing anemia. The same was true in cells from DBA patients transplanted into mice.
The higher the dose of SMER28, the more RBCs were produced, and no ill effects were found. However, formal toxicity studies have not been conducted.
SMER28 has been tested preclinically for some neurodegenerative diseases. It activates an autophagy pathway that recycles damaged cellular components.
In DBA, SMER28 appears to turn on autophagy in erythroid progenitors. Dr Doulatov plans to further explore how this interferes with RBC production. ![]()
Resistant malaria parasite spreading through Asia
Image courtesy of
CDC/Mae Melvin
A lineage of multidrug-resistant malaria parasite has spread widely and is now established in parts of Thailand, Laos, and Cambodia, according to a study published in The Lancet Infectious Diseases.
The
researchers said their findings suggest an artemisinin-resistant
Plasmodium falciparum lineage probably arose in western Cambodia and
then spread to Thailand and Laos, outcompeting other parasites and
acquiring resistance to piperaquine.
“We now see this very successful, resistant parasite lineage emerging, outcompeting its peers, and spreading over a wide area,” said study author Arjen Dondorp, MD, of Mahidol University in Bangkok, Thailand, and the University of Oxford in Oxford, UK.
“It has also picked up resistance to the partner drug piperaquine, causing high failure rates of the widely used artemisinin combination therapy, DHA-piperaquine. We hope this evidence will be used to re-emphasize the urgency of malaria elimination in the Asia region before falciparum malaria becomes close to untreatable.”
For this study, Dr Dondorp and his colleagues examined blood spot samples from patients with uncomplicated, P falciparum malaria collected at sites in Cambodia, Laos, Thailand, and Myanmar.
The researchers found evidence to suggest that PfKelch13 C580Y, a single mutant parasite lineage, has spread across Cambodia, Laos, and Thailand, replacing parasites containing other, less resistant mutations.
Although the C580Y mutation does not confer a higher level of artemisinin resistance than many other PfKelch13 mutations, it appears to be fitter, more transmissible, and spreading more widely.
“It isn’t that the C580Y mutation itself makes the malaria parasites fitter, it is the other genetic changes that go along with it—hence, the critical emphasis on the term ‘lineage,’” said Nicholas White, also of Mahidol University and the University of Oxford.
“This is what makes superbugs—the evolution of multiple factors that make them fitter and more transmissible. The spread and emergence of drug-resistant malaria parasites across Asia into Africa has occurred before. Last time, it killed millions. We need to work with our policy, research, and funding partners to respond to this threat in Asia urgently to avoid history repeating itself.”
In particular, the researchers are calling for accelerated efforts in the Greater Mekong subregion and closer collaboration to monitor any further spread in neighboring regions.
“We are losing a dangerous race to eliminate artemisinin-resistant falciparum malaria before widespread resistance to the partner antimalarials makes that impossible,” White said. “The consequences of resistance spreading further into India and Africa could be grave if drug resistance is not tackled from a global public health emergency perspective.” ![]()

Image courtesy of
CDC/Mae Melvin
A lineage of multidrug-resistant malaria parasite has spread widely and is now established in parts of Thailand, Laos, and Cambodia, according to a study published in The Lancet Infectious Diseases.
The
researchers said their findings suggest an artemisinin-resistant
Plasmodium falciparum lineage probably arose in western Cambodia and
then spread to Thailand and Laos, outcompeting other parasites and
acquiring resistance to piperaquine.
“We now see this very successful, resistant parasite lineage emerging, outcompeting its peers, and spreading over a wide area,” said study author Arjen Dondorp, MD, of Mahidol University in Bangkok, Thailand, and the University of Oxford in Oxford, UK.
“It has also picked up resistance to the partner drug piperaquine, causing high failure rates of the widely used artemisinin combination therapy, DHA-piperaquine. We hope this evidence will be used to re-emphasize the urgency of malaria elimination in the Asia region before falciparum malaria becomes close to untreatable.”
For this study, Dr Dondorp and his colleagues examined blood spot samples from patients with uncomplicated, P falciparum malaria collected at sites in Cambodia, Laos, Thailand, and Myanmar.
The researchers found evidence to suggest that PfKelch13 C580Y, a single mutant parasite lineage, has spread across Cambodia, Laos, and Thailand, replacing parasites containing other, less resistant mutations.
Although the C580Y mutation does not confer a higher level of artemisinin resistance than many other PfKelch13 mutations, it appears to be fitter, more transmissible, and spreading more widely.
“It isn’t that the C580Y mutation itself makes the malaria parasites fitter, it is the other genetic changes that go along with it—hence, the critical emphasis on the term ‘lineage,’” said Nicholas White, also of Mahidol University and the University of Oxford.
“This is what makes superbugs—the evolution of multiple factors that make them fitter and more transmissible. The spread and emergence of drug-resistant malaria parasites across Asia into Africa has occurred before. Last time, it killed millions. We need to work with our policy, research, and funding partners to respond to this threat in Asia urgently to avoid history repeating itself.”
In particular, the researchers are calling for accelerated efforts in the Greater Mekong subregion and closer collaboration to monitor any further spread in neighboring regions.
“We are losing a dangerous race to eliminate artemisinin-resistant falciparum malaria before widespread resistance to the partner antimalarials makes that impossible,” White said. “The consequences of resistance spreading further into India and Africa could be grave if drug resistance is not tackled from a global public health emergency perspective.” ![]()

Image courtesy of
CDC/Mae Melvin
A lineage of multidrug-resistant malaria parasite has spread widely and is now established in parts of Thailand, Laos, and Cambodia, according to a study published in The Lancet Infectious Diseases.
The
researchers said their findings suggest an artemisinin-resistant
Plasmodium falciparum lineage probably arose in western Cambodia and
then spread to Thailand and Laos, outcompeting other parasites and
acquiring resistance to piperaquine.
“We now see this very successful, resistant parasite lineage emerging, outcompeting its peers, and spreading over a wide area,” said study author Arjen Dondorp, MD, of Mahidol University in Bangkok, Thailand, and the University of Oxford in Oxford, UK.
“It has also picked up resistance to the partner drug piperaquine, causing high failure rates of the widely used artemisinin combination therapy, DHA-piperaquine. We hope this evidence will be used to re-emphasize the urgency of malaria elimination in the Asia region before falciparum malaria becomes close to untreatable.”
For this study, Dr Dondorp and his colleagues examined blood spot samples from patients with uncomplicated, P falciparum malaria collected at sites in Cambodia, Laos, Thailand, and Myanmar.
The researchers found evidence to suggest that PfKelch13 C580Y, a single mutant parasite lineage, has spread across Cambodia, Laos, and Thailand, replacing parasites containing other, less resistant mutations.
Although the C580Y mutation does not confer a higher level of artemisinin resistance than many other PfKelch13 mutations, it appears to be fitter, more transmissible, and spreading more widely.
“It isn’t that the C580Y mutation itself makes the malaria parasites fitter, it is the other genetic changes that go along with it—hence, the critical emphasis on the term ‘lineage,’” said Nicholas White, also of Mahidol University and the University of Oxford.
“This is what makes superbugs—the evolution of multiple factors that make them fitter and more transmissible. The spread and emergence of drug-resistant malaria parasites across Asia into Africa has occurred before. Last time, it killed millions. We need to work with our policy, research, and funding partners to respond to this threat in Asia urgently to avoid history repeating itself.”
In particular, the researchers are calling for accelerated efforts in the Greater Mekong subregion and closer collaboration to monitor any further spread in neighboring regions.
“We are losing a dangerous race to eliminate artemisinin-resistant falciparum malaria before widespread resistance to the partner antimalarials makes that impossible,” White said. “The consequences of resistance spreading further into India and Africa could be grave if drug resistance is not tackled from a global public health emergency perspective.”
FDA approves IVIG product for PI and chronic ITP

Photo by Bill Branson
The US Food and Drug Administration (FDA) has approved an intravenous immunoglobulin (IVIG) product (Gammaplex® 10%) for the treatment of primary immunodeficiency (PI) and chronic immune thrombocytopenia (ITP) in adults.
PI includes, but is not limited to,
the humoral immune defect in common variable immunodeficiency, X-linked and congenital
agammaglobulinemia, Wiskott-Aldrich
syndrome, and severe combined immunodeficiencies.
Gammaplex 10% is manufactured by Bio Products Laboratory Limited (BPL).
Gammaplex 10% is made with the same process as BPL’s previously approved IVIG treatment, Gammaplex® 5% (immune globulin intravenous [human], 5% liquid).
Gammaplex 10% is more concentrated than Gammaplex 5%, with an immune globulin G (IgG) concentration of 100 g/L, and is stabilized with glycine.
The FDA’s approval of Gammaplex 10% was based on a 2-phase, crossover bioequivalence study comparing Gammaplex 10% and Gammaplex 5% in 33 adult patients with PI. This study is the first direct comparison of 10% and 5% IVIG products in the treatment of PI.
The primary endpoint of bioequivalence between the products was achieved, and trough levels of IgG were well maintained throughout the study.
Both Gammaplex 10% and Gammaplex 5% infusion rates were increased incrementally at 15-minute intervals if tolerated by the subject. The Gammaplex 10% infusion rate was increased to the maximum in 96% of infusions.
The mean infusion time for Gammaplex 10% was 1 hour and 51 minutes, which was 57 minutes faster than Gammaplex 5%.
There were no notable differences in the safety and tolerability of the 2 products.
The most common adverse events in patients receiving Gammaplex 10% were headache (12.5%), migraine (6.3%), and pyrexia (6.3%). There were no serious product-related adverse events.
The safety of Gammaplex 10% has not been established in adults with chronic ITP. The safety profile for Gammaplex 5% has been studied in a phase 3 trial of adults with chronic ITP, and it is anticipated that the safety profile for both formulations are comparable for ITP patients.
The most common adverse events in adults with chronic ITP receiving Gammaplex 5% were headache, vomiting, nausea, pyrexia, arthralgia, and dehydration. Serious adverse events were headache, vomiting, and dehydration.
The full prescribing information for Gammaplex 10%, which includes trial data, is available at http://www.gammaplex.com.

Photo by Bill Branson
The US Food and Drug Administration (FDA) has approved an intravenous immunoglobulin (IVIG) product (Gammaplex® 10%) for the treatment of primary immunodeficiency (PI) and chronic immune thrombocytopenia (ITP) in adults.
PI includes, but is not limited to,
the humoral immune defect in common variable immunodeficiency, X-linked and congenital
agammaglobulinemia, Wiskott-Aldrich
syndrome, and severe combined immunodeficiencies.
Gammaplex 10% is manufactured by Bio Products Laboratory Limited (BPL).
Gammaplex 10% is made with the same process as BPL’s previously approved IVIG treatment, Gammaplex® 5% (immune globulin intravenous [human], 5% liquid).
Gammaplex 10% is more concentrated than Gammaplex 5%, with an immune globulin G (IgG) concentration of 100 g/L, and is stabilized with glycine.
The FDA’s approval of Gammaplex 10% was based on a 2-phase, crossover bioequivalence study comparing Gammaplex 10% and Gammaplex 5% in 33 adult patients with PI. This study is the first direct comparison of 10% and 5% IVIG products in the treatment of PI.
The primary endpoint of bioequivalence between the products was achieved, and trough levels of IgG were well maintained throughout the study.
Both Gammaplex 10% and Gammaplex 5% infusion rates were increased incrementally at 15-minute intervals if tolerated by the subject. The Gammaplex 10% infusion rate was increased to the maximum in 96% of infusions.
The mean infusion time for Gammaplex 10% was 1 hour and 51 minutes, which was 57 minutes faster than Gammaplex 5%.
There were no notable differences in the safety and tolerability of the 2 products.
The most common adverse events in patients receiving Gammaplex 10% were headache (12.5%), migraine (6.3%), and pyrexia (6.3%). There were no serious product-related adverse events.
The safety of Gammaplex 10% has not been established in adults with chronic ITP. The safety profile for Gammaplex 5% has been studied in a phase 3 trial of adults with chronic ITP, and it is anticipated that the safety profile for both formulations are comparable for ITP patients.
The most common adverse events in adults with chronic ITP receiving Gammaplex 5% were headache, vomiting, nausea, pyrexia, arthralgia, and dehydration. Serious adverse events were headache, vomiting, and dehydration.
The full prescribing information for Gammaplex 10%, which includes trial data, is available at http://www.gammaplex.com.

Photo by Bill Branson
The US Food and Drug Administration (FDA) has approved an intravenous immunoglobulin (IVIG) product (Gammaplex® 10%) for the treatment of primary immunodeficiency (PI) and chronic immune thrombocytopenia (ITP) in adults.
PI includes, but is not limited to,
the humoral immune defect in common variable immunodeficiency, X-linked and congenital
agammaglobulinemia, Wiskott-Aldrich
syndrome, and severe combined immunodeficiencies.
Gammaplex 10% is manufactured by Bio Products Laboratory Limited (BPL).
Gammaplex 10% is made with the same process as BPL’s previously approved IVIG treatment, Gammaplex® 5% (immune globulin intravenous [human], 5% liquid).
Gammaplex 10% is more concentrated than Gammaplex 5%, with an immune globulin G (IgG) concentration of 100 g/L, and is stabilized with glycine.
The FDA’s approval of Gammaplex 10% was based on a 2-phase, crossover bioequivalence study comparing Gammaplex 10% and Gammaplex 5% in 33 adult patients with PI. This study is the first direct comparison of 10% and 5% IVIG products in the treatment of PI.
The primary endpoint of bioequivalence between the products was achieved, and trough levels of IgG were well maintained throughout the study.
Both Gammaplex 10% and Gammaplex 5% infusion rates were increased incrementally at 15-minute intervals if tolerated by the subject. The Gammaplex 10% infusion rate was increased to the maximum in 96% of infusions.
The mean infusion time for Gammaplex 10% was 1 hour and 51 minutes, which was 57 minutes faster than Gammaplex 5%.
There were no notable differences in the safety and tolerability of the 2 products.
The most common adverse events in patients receiving Gammaplex 10% were headache (12.5%), migraine (6.3%), and pyrexia (6.3%). There were no serious product-related adverse events.
The safety of Gammaplex 10% has not been established in adults with chronic ITP. The safety profile for Gammaplex 5% has been studied in a phase 3 trial of adults with chronic ITP, and it is anticipated that the safety profile for both formulations are comparable for ITP patients.
The most common adverse events in adults with chronic ITP receiving Gammaplex 5% were headache, vomiting, nausea, pyrexia, arthralgia, and dehydration. Serious adverse events were headache, vomiting, and dehydration.
The full prescribing information for Gammaplex 10%, which includes trial data, is available at http://www.gammaplex.com.
Investigators report new risk loci for CLL
Investigators say they have identified 9 new risk loci for chronic lymphocytic leukemia (CLL).
The team says the research, published in Nature Communications, provides additional evidence for genetic susceptibility to CLL and sheds new light on the biological basis of CLL development.
They also believe their findings could aid the development of new drugs for CLL or help in selecting existing therapies for CLL
patients.
“We knew people were more likely to develop chronic lymphocytic leukemia if someone in their family had suffered from the disease, but our new research takes a big step towards explaining the underlying genetics,” said study author Richard Houlston, MD, PhD, of The Institute of Cancer Research in London, UK.
“CLL is essentially a disease of the immune system, and it’s fascinating that so many of the new genetic variants we have uncovered seem to directly affect the behavior of white blood cells and their ability to fight disease. Understanding the genetics of CLL can point us towards new treatments for the disease and help us to use existing targeted drugs more effectively.”
For this study, Dr Houlston and his colleagues analyzed data from 8 studies involving a total of 6200 CLL patients and 17,598 controls.
From this, the team identified 9 CLL risk loci:
- 1p36.11 (rs34676223, P=5.04 × 10−13)
- 1q42.13 (rs41271473, P=1.06 × 10−10)
- 4q24 (rs71597109, P=1.37 × 10−10)
- 4q35.1 (rs57214277, P=3.69 × 10−8)
- 6p21.31 (rs3800461, P=1.97 × 10−8)
- 11q23.2 (rs61904987, P=2.64 × 10−11)
- 18q21.1 (rs1036935, P=3.27 × 10−8)
- 19p13.3 (rs7254272, P=4.67 × 10−8)
- 22q13.33 (rs140522, P=2.70 × 10−9).
The investigators noted that the 4q24 association marked by rs71597109 maps to intron 1 of the gene encoding BANK1 (B-cell scaffold protein with ankyrin repeats 1). BANK1 is only ever activated in B cells and is linked to the autoimmune disease lupus.
The team also pointed out that the 19p13.3 association marked by rs7254272 maps 2.5 kb 5′ to ZBTB7A (zinc finger and BTB domain-containing protein 7a), which is a master regulator of B versus T lymphoid fate. So errors in ZBTB7A could lead to too many B cells in the bloodstream and bone marrow.
And rs140522 maps to 22q13.33, which has been linked to the development of multiple sclerosis. The investigators noted that this region of linkage disequilibrium contains 4 genes. One of them, NCAPH2 (non-SMC condensin II complex subunit H2), is differentially expressed in CLL and normal B cells.
“This fascinating study makes a link between genetic variants in the immune system and the development of leukemia and implicates regions of DNA which are also involved in autoimmune diseases,” said Paul Workman, PhD, chief executive and president of The Institute of Cancer Research, who was not involved in this research.
“The findings could point us towards new ways of treating leukemia or better ways of using existing treatments—potentially including immunotherapies.”

Investigators say they have identified 9 new risk loci for chronic lymphocytic leukemia (CLL).
The team says the research, published in Nature Communications, provides additional evidence for genetic susceptibility to CLL and sheds new light on the biological basis of CLL development.
They also believe their findings could aid the development of new drugs for CLL or help in selecting existing therapies for CLL
patients.
“We knew people were more likely to develop chronic lymphocytic leukemia if someone in their family had suffered from the disease, but our new research takes a big step towards explaining the underlying genetics,” said study author Richard Houlston, MD, PhD, of The Institute of Cancer Research in London, UK.
“CLL is essentially a disease of the immune system, and it’s fascinating that so many of the new genetic variants we have uncovered seem to directly affect the behavior of white blood cells and their ability to fight disease. Understanding the genetics of CLL can point us towards new treatments for the disease and help us to use existing targeted drugs more effectively.”
For this study, Dr Houlston and his colleagues analyzed data from 8 studies involving a total of 6200 CLL patients and 17,598 controls.
From this, the team identified 9 CLL risk loci:
- 1p36.11 (rs34676223, P=5.04 × 10−13)
- 1q42.13 (rs41271473, P=1.06 × 10−10)
- 4q24 (rs71597109, P=1.37 × 10−10)
- 4q35.1 (rs57214277, P=3.69 × 10−8)
- 6p21.31 (rs3800461, P=1.97 × 10−8)
- 11q23.2 (rs61904987, P=2.64 × 10−11)
- 18q21.1 (rs1036935, P=3.27 × 10−8)
- 19p13.3 (rs7254272, P=4.67 × 10−8)
- 22q13.33 (rs140522, P=2.70 × 10−9).
The investigators noted that the 4q24 association marked by rs71597109 maps to intron 1 of the gene encoding BANK1 (B-cell scaffold protein with ankyrin repeats 1). BANK1 is only ever activated in B cells and is linked to the autoimmune disease lupus.
The team also pointed out that the 19p13.3 association marked by rs7254272 maps 2.5 kb 5′ to ZBTB7A (zinc finger and BTB domain-containing protein 7a), which is a master regulator of B versus T lymphoid fate. So errors in ZBTB7A could lead to too many B cells in the bloodstream and bone marrow.
And rs140522 maps to 22q13.33, which has been linked to the development of multiple sclerosis. The investigators noted that this region of linkage disequilibrium contains 4 genes. One of them, NCAPH2 (non-SMC condensin II complex subunit H2), is differentially expressed in CLL and normal B cells.
“This fascinating study makes a link between genetic variants in the immune system and the development of leukemia and implicates regions of DNA which are also involved in autoimmune diseases,” said Paul Workman, PhD, chief executive and president of The Institute of Cancer Research, who was not involved in this research.
“The findings could point us towards new ways of treating leukemia or better ways of using existing treatments—potentially including immunotherapies.”

Investigators say they have identified 9 new risk loci for chronic lymphocytic leukemia (CLL).
The team says the research, published in Nature Communications, provides additional evidence for genetic susceptibility to CLL and sheds new light on the biological basis of CLL development.
They also believe their findings could aid the development of new drugs for CLL or help in selecting existing therapies for CLL
patients.
“We knew people were more likely to develop chronic lymphocytic leukemia if someone in their family had suffered from the disease, but our new research takes a big step towards explaining the underlying genetics,” said study author Richard Houlston, MD, PhD, of The Institute of Cancer Research in London, UK.
“CLL is essentially a disease of the immune system, and it’s fascinating that so many of the new genetic variants we have uncovered seem to directly affect the behavior of white blood cells and their ability to fight disease. Understanding the genetics of CLL can point us towards new treatments for the disease and help us to use existing targeted drugs more effectively.”
For this study, Dr Houlston and his colleagues analyzed data from 8 studies involving a total of 6200 CLL patients and 17,598 controls.
From this, the team identified 9 CLL risk loci:
- 1p36.11 (rs34676223, P=5.04 × 10−13)
- 1q42.13 (rs41271473, P=1.06 × 10−10)
- 4q24 (rs71597109, P=1.37 × 10−10)
- 4q35.1 (rs57214277, P=3.69 × 10−8)
- 6p21.31 (rs3800461, P=1.97 × 10−8)
- 11q23.2 (rs61904987, P=2.64 × 10−11)
- 18q21.1 (rs1036935, P=3.27 × 10−8)
- 19p13.3 (rs7254272, P=4.67 × 10−8)
- 22q13.33 (rs140522, P=2.70 × 10−9).
The investigators noted that the 4q24 association marked by rs71597109 maps to intron 1 of the gene encoding BANK1 (B-cell scaffold protein with ankyrin repeats 1). BANK1 is only ever activated in B cells and is linked to the autoimmune disease lupus.
The team also pointed out that the 19p13.3 association marked by rs7254272 maps 2.5 kb 5′ to ZBTB7A (zinc finger and BTB domain-containing protein 7a), which is a master regulator of B versus T lymphoid fate. So errors in ZBTB7A could lead to too many B cells in the bloodstream and bone marrow.
And rs140522 maps to 22q13.33, which has been linked to the development of multiple sclerosis. The investigators noted that this region of linkage disequilibrium contains 4 genes. One of them, NCAPH2 (non-SMC condensin II complex subunit H2), is differentially expressed in CLL and normal B cells.
“This fascinating study makes a link between genetic variants in the immune system and the development of leukemia and implicates regions of DNA which are also involved in autoimmune diseases,” said Paul Workman, PhD, chief executive and president of The Institute of Cancer Research, who was not involved in this research.
“The findings could point us towards new ways of treating leukemia or better ways of using existing treatments—potentially including immunotherapies.”
Obinutuzumab approved to treat FL in Canada

Health Canada has approved the use of obinutuzumab (Gazyva®), an anti-CD20 monoclonal antibody, in patients with follicular lymphoma (FL).
The approval means obinutuzumab can be given, first in combination with bendamustine and then alone as maintenance therapy, to FL patients who relapsed after, or are refractory to, a rituximab-containing regimen.
Obinutuzumab is also approved in Canada for use in combination with chlorambucil to treat patients with previously untreated chronic lymphocytic leukemia.
Obinutuzumab is a product of Roche.
Health Canada’s approval of obinutuzumab in FL is based on results from the phase 3 GADOLIN trial.
The study included 413 patients with rituximab-refractory non-Hodgkin lymphoma, including 321 patients with FL, 46 with marginal zone lymphoma, and 28 with small lymphocytic lymphoma.
The patients were randomized to receive bendamustine alone (control arm) or a combination of bendamustine and obinutuzumab followed by obinutuzumab maintenance (every 2 months for 2 years or until progression).
The primary endpoint of the study was progression-free survival (PFS), as assessed by an independent review committee (IRC). The secondary endpoints were PFS assessed by investigator review, best overall response, complete response (CR), partial response (PR), duration of response, overall survival, and safety profile.
Among patients with FL, the obinutuzumab regimen improved PFS compared to bendamustine alone, as assessed by the IRC (hazard ratio [HR]=0.48, P<0.0001). The median PFS was not reached in patients receiving the obinutuzumab regimen but was 13.8 months in those receiving bendamustine alone.
Investigator-assessed PFS was consistent with IRC-assessed PFS. Investigators said the median PFS with the obinutuzumab regimen was more than double that with bendamustine alone—29.2 months vs 13.7 months (HR=0.48, P<0.0001).
The best overall response for patients receiving the obinutuzumab regimen was 78.7% (15.5% CR, 63.2% PR), compared to 74.7% (18.7% CR, 56% PR) for those receiving bendamustine alone, as assessed by the IRC.
The median duration of response was not reached for patients receiving the obinutuzumab regimen and was 11.6 months for those receiving bendamustine alone.
At last follow-up, the median overall survival had not been reached in either study arm.
The most common grade 3/4 adverse events observed in patients receiving the obinutuzumab regimen were neutropenia (33%), infusion reactions (11%), and thrombocytopenia (10%).
The most common adverse events of any grade were infusion reactions (69%), neutropenia (35%), nausea (54%), fatigue (39%), cough (26%), diarrhea (27%), constipation (19%), fever (18%), thrombocytopenia (15%), vomiting (22%), upper respiratory tract infection (13%), decreased appetite (18%), joint or muscle pain (12%), sinusitis (12%), anemia (12%), general weakness (11%), and urinary tract infection (10%).

Health Canada has approved the use of obinutuzumab (Gazyva®), an anti-CD20 monoclonal antibody, in patients with follicular lymphoma (FL).
The approval means obinutuzumab can be given, first in combination with bendamustine and then alone as maintenance therapy, to FL patients who relapsed after, or are refractory to, a rituximab-containing regimen.
Obinutuzumab is also approved in Canada for use in combination with chlorambucil to treat patients with previously untreated chronic lymphocytic leukemia.
Obinutuzumab is a product of Roche.
Health Canada’s approval of obinutuzumab in FL is based on results from the phase 3 GADOLIN trial.
The study included 413 patients with rituximab-refractory non-Hodgkin lymphoma, including 321 patients with FL, 46 with marginal zone lymphoma, and 28 with small lymphocytic lymphoma.
The patients were randomized to receive bendamustine alone (control arm) or a combination of bendamustine and obinutuzumab followed by obinutuzumab maintenance (every 2 months for 2 years or until progression).
The primary endpoint of the study was progression-free survival (PFS), as assessed by an independent review committee (IRC). The secondary endpoints were PFS assessed by investigator review, best overall response, complete response (CR), partial response (PR), duration of response, overall survival, and safety profile.
Among patients with FL, the obinutuzumab regimen improved PFS compared to bendamustine alone, as assessed by the IRC (hazard ratio [HR]=0.48, P<0.0001). The median PFS was not reached in patients receiving the obinutuzumab regimen but was 13.8 months in those receiving bendamustine alone.
Investigator-assessed PFS was consistent with IRC-assessed PFS. Investigators said the median PFS with the obinutuzumab regimen was more than double that with bendamustine alone—29.2 months vs 13.7 months (HR=0.48, P<0.0001).
The best overall response for patients receiving the obinutuzumab regimen was 78.7% (15.5% CR, 63.2% PR), compared to 74.7% (18.7% CR, 56% PR) for those receiving bendamustine alone, as assessed by the IRC.
The median duration of response was not reached for patients receiving the obinutuzumab regimen and was 11.6 months for those receiving bendamustine alone.
At last follow-up, the median overall survival had not been reached in either study arm.
The most common grade 3/4 adverse events observed in patients receiving the obinutuzumab regimen were neutropenia (33%), infusion reactions (11%), and thrombocytopenia (10%).
The most common adverse events of any grade were infusion reactions (69%), neutropenia (35%), nausea (54%), fatigue (39%), cough (26%), diarrhea (27%), constipation (19%), fever (18%), thrombocytopenia (15%), vomiting (22%), upper respiratory tract infection (13%), decreased appetite (18%), joint or muscle pain (12%), sinusitis (12%), anemia (12%), general weakness (11%), and urinary tract infection (10%).

Health Canada has approved the use of obinutuzumab (Gazyva®), an anti-CD20 monoclonal antibody, in patients with follicular lymphoma (FL).
The approval means obinutuzumab can be given, first in combination with bendamustine and then alone as maintenance therapy, to FL patients who relapsed after, or are refractory to, a rituximab-containing regimen.
Obinutuzumab is also approved in Canada for use in combination with chlorambucil to treat patients with previously untreated chronic lymphocytic leukemia.
Obinutuzumab is a product of Roche.
Health Canada’s approval of obinutuzumab in FL is based on results from the phase 3 GADOLIN trial.
The study included 413 patients with rituximab-refractory non-Hodgkin lymphoma, including 321 patients with FL, 46 with marginal zone lymphoma, and 28 with small lymphocytic lymphoma.
The patients were randomized to receive bendamustine alone (control arm) or a combination of bendamustine and obinutuzumab followed by obinutuzumab maintenance (every 2 months for 2 years or until progression).
The primary endpoint of the study was progression-free survival (PFS), as assessed by an independent review committee (IRC). The secondary endpoints were PFS assessed by investigator review, best overall response, complete response (CR), partial response (PR), duration of response, overall survival, and safety profile.
Among patients with FL, the obinutuzumab regimen improved PFS compared to bendamustine alone, as assessed by the IRC (hazard ratio [HR]=0.48, P<0.0001). The median PFS was not reached in patients receiving the obinutuzumab regimen but was 13.8 months in those receiving bendamustine alone.
Investigator-assessed PFS was consistent with IRC-assessed PFS. Investigators said the median PFS with the obinutuzumab regimen was more than double that with bendamustine alone—29.2 months vs 13.7 months (HR=0.48, P<0.0001).
The best overall response for patients receiving the obinutuzumab regimen was 78.7% (15.5% CR, 63.2% PR), compared to 74.7% (18.7% CR, 56% PR) for those receiving bendamustine alone, as assessed by the IRC.
The median duration of response was not reached for patients receiving the obinutuzumab regimen and was 11.6 months for those receiving bendamustine alone.
At last follow-up, the median overall survival had not been reached in either study arm.
The most common grade 3/4 adverse events observed in patients receiving the obinutuzumab regimen were neutropenia (33%), infusion reactions (11%), and thrombocytopenia (10%).
The most common adverse events of any grade were infusion reactions (69%), neutropenia (35%), nausea (54%), fatigue (39%), cough (26%), diarrhea (27%), constipation (19%), fever (18%), thrombocytopenia (15%), vomiting (22%), upper respiratory tract infection (13%), decreased appetite (18%), joint or muscle pain (12%), sinusitis (12%), anemia (12%), general weakness (11%), and urinary tract infection (10%).
