User login
FDA approves new formulation of mercaptopurine
Credit: Bill Branson
The US Food and Drug Administration (FDA) has approved an oral suspension of mercaptopurine (Purixan), a drug used in combination therapy to treat patients with acute lymphoblastic leukemia (ALL).
Mercaptopurine will now be available as a 20 mg/mL oral suspension.
The drug was originally approved as a 50 mg tablet in 1953 and, since that time, has only been commercially available in this form.
Unfortunately, a 50 mg tablet is not ideal because of the age and weight range of children with ALL.
The tablet does not allow for easy body-surface-area dosing and dose adjustments. In addition, tablets can be difficult to administer to children younger than 6 years of age.
To overcome these issues, physicians have used ad hoc local formulations of mercaptopurine compounded in pharmacies or recommended splitting tablets to provide children with the desired dose.
According to the FDA, offering mercaptopurine as a suspension will allow for more accurate delivery of the desired dose to children with a wide range of weights. And a commercially produced suspension is likely to provide a more consistent dose of 6-mercaptopurine than ad hoc compounded formulations.
The FDA’s approval of mercaptopurine as a 20 mg/mL oral suspension is based on results of a clinical pharmacology study. The goal of the study was to assess the bioequivalence of mercaptopurine from a tablet with that of the mercaptopurine oral suspension in a healthy adult population.
The starting dose of mercaptopurine in multi-agent combination chemotherapy maintenance regimens is 1.5 mg/kg to 2.5 mg/kg (50 mg/m2 to 75 mg/m2) as a single, daily dose.
After initiating mercaptopurine, continuation of appropriate dosing requires periodic monitoring of absolute neutrophil counts and platelet counts to assure sufficient drug exposure and to adjust for excessive hematologic toxicity.
Mercaptopurine is distributed by Rare Disease Therapeutics, Inc., in Nashville, Tennessee. For more details on the drug, see the prescribing information. ![]()

Credit: Bill Branson
The US Food and Drug Administration (FDA) has approved an oral suspension of mercaptopurine (Purixan), a drug used in combination therapy to treat patients with acute lymphoblastic leukemia (ALL).
Mercaptopurine will now be available as a 20 mg/mL oral suspension.
The drug was originally approved as a 50 mg tablet in 1953 and, since that time, has only been commercially available in this form.
Unfortunately, a 50 mg tablet is not ideal because of the age and weight range of children with ALL.
The tablet does not allow for easy body-surface-area dosing and dose adjustments. In addition, tablets can be difficult to administer to children younger than 6 years of age.
To overcome these issues, physicians have used ad hoc local formulations of mercaptopurine compounded in pharmacies or recommended splitting tablets to provide children with the desired dose.
According to the FDA, offering mercaptopurine as a suspension will allow for more accurate delivery of the desired dose to children with a wide range of weights. And a commercially produced suspension is likely to provide a more consistent dose of 6-mercaptopurine than ad hoc compounded formulations.
The FDA’s approval of mercaptopurine as a 20 mg/mL oral suspension is based on results of a clinical pharmacology study. The goal of the study was to assess the bioequivalence of mercaptopurine from a tablet with that of the mercaptopurine oral suspension in a healthy adult population.
The starting dose of mercaptopurine in multi-agent combination chemotherapy maintenance regimens is 1.5 mg/kg to 2.5 mg/kg (50 mg/m2 to 75 mg/m2) as a single, daily dose.
After initiating mercaptopurine, continuation of appropriate dosing requires periodic monitoring of absolute neutrophil counts and platelet counts to assure sufficient drug exposure and to adjust for excessive hematologic toxicity.
Mercaptopurine is distributed by Rare Disease Therapeutics, Inc., in Nashville, Tennessee. For more details on the drug, see the prescribing information. ![]()

Credit: Bill Branson
The US Food and Drug Administration (FDA) has approved an oral suspension of mercaptopurine (Purixan), a drug used in combination therapy to treat patients with acute lymphoblastic leukemia (ALL).
Mercaptopurine will now be available as a 20 mg/mL oral suspension.
The drug was originally approved as a 50 mg tablet in 1953 and, since that time, has only been commercially available in this form.
Unfortunately, a 50 mg tablet is not ideal because of the age and weight range of children with ALL.
The tablet does not allow for easy body-surface-area dosing and dose adjustments. In addition, tablets can be difficult to administer to children younger than 6 years of age.
To overcome these issues, physicians have used ad hoc local formulations of mercaptopurine compounded in pharmacies or recommended splitting tablets to provide children with the desired dose.
According to the FDA, offering mercaptopurine as a suspension will allow for more accurate delivery of the desired dose to children with a wide range of weights. And a commercially produced suspension is likely to provide a more consistent dose of 6-mercaptopurine than ad hoc compounded formulations.
The FDA’s approval of mercaptopurine as a 20 mg/mL oral suspension is based on results of a clinical pharmacology study. The goal of the study was to assess the bioequivalence of mercaptopurine from a tablet with that of the mercaptopurine oral suspension in a healthy adult population.
The starting dose of mercaptopurine in multi-agent combination chemotherapy maintenance regimens is 1.5 mg/kg to 2.5 mg/kg (50 mg/m2 to 75 mg/m2) as a single, daily dose.
After initiating mercaptopurine, continuation of appropriate dosing requires periodic monitoring of absolute neutrophil counts and platelet counts to assure sufficient drug exposure and to adjust for excessive hematologic toxicity.
Mercaptopurine is distributed by Rare Disease Therapeutics, Inc., in Nashville, Tennessee. For more details on the drug, see the prescribing information. ![]()
Leukemic breast tumors may cause resistance in AML, ALL
SAN DIEGO—One woman’s curiosity and self-described “aggressive” approach to research have led to some unexpected discoveries about acute leukemias.
Isabel Cunningham, MD, of Columbia University in New York, has found evidence to suggest that treatment resistance in leukemia patients may sometimes result from an interaction between leukemic cells and the breast.
She discovered that leukemic cells in extramedullary niches can adopt a tumor phenotype similar to breast cancer.
And many genes are similarly upregulated in leukemic and epithelial breast tumors.
Her research indicates that a new approach to resistant leukemias that incorporates the principles of solid-tumor treatment—scans to identify any tumors and surgery to remove them—could decrease marrow relapse and death.
Dr Cunningham and her colleagues presented these findings in a poster at the AACR Annual Meeting 2014 (abstract 3996*).
“Chemotherapy resistance is our main problem in treating leukemia,” Dr Cunningham said. “It’s been known for a long time that, occasionally, leukemia forms tumors in an organ, but there’s never been a unified approach to treatment, except for leukemia that occurs in the testis and the meninges.”
Dr Cunningham had encountered many patients with resistant leukemia throughout her career, but her research actually began with a patient she had never met. A case study of a leukemia patient with a breast tumor sparked Dr Cunningham’s interest, and she emailed the study’s author to find out what ultimately became of the patient.
The response she received peaked her curiosity further. So she began seeking more of these cases, contacting authors, and collecting information on this phenomenon.
“I took this on as sort of a hobby,” Dr Cunningham said. “I never had any idea where this was going to lead.”
Eventually, she had amassed information on 235 cases—163 patients with acute myeloid leukemia (AML) and 72 with acute lymphoblastic leukemia (ALL)—who ranged from 1 year to 75 years of age. And an analysis of these cases led to some surprising discoveries.
Clinical findings
Dr Cunningham found these leukemic breast tumors can occur before, during, or after marrow leukemia. And, clinically, they resemble breast cancer. Most tumors were palpable, and some were detected only on routine mammograms.
There were single or multiple nodules that may have involved the entire breast. Sixty percent of cases were unilateral on presentation, but, often, the other breast became involved. Seventy percent of cases exhibited axillary lymphadenopathy that was ipsilateral.
Most tumors grew rapidly, to as large as 12 cm. The tumor behavior was similar in AML and ALL. And the tumors had a metastatic pattern similar to lobular breast cancer—spreading to the contralateral breast, the abdomen or pelvis, the meninges, and culminating in death.
However, some patients did survive. Four percent of patients who were treated only with chemotherapy were alive at 4 years. Twenty-five percent of patients had their tumors excised prior to chemotherapy and were alive anywhere from 3 years to more than 26 years after treatment.
Histology and gene expression
To build upon these findings, Dr Cunningham set her sights on patient samples. She was able to obtain paraffin blocks of leukemic breast tumors from 25 patients and perform immunohistochemical staining.
“It became clear that the leukemic tumors—which are marked by leukemic markers and not breast cancer markers—look, histologically, like breast cancer, specifically, lobular breast cancer,” Dr Cunningham said. “An additional pathologic finding was a specific type of desmoplastic fibrosis seen in all 25 contributed biopsies.”
Dr Cunningham also performed gene expression studies on 3 of the tumors (2 ALL and 1 AML), which were collected 8 months to 22 months after diagnosis, while marrows were in remission. The analyses revealed that a number of genes are significantly upregulated in both leukemic breast tumors and breast cancer.
These include genes involved in adhesion and interactions with the extracellular matrix (ADAM8, COMP, and CDH22), genes involved in the ubiquitin-proteasome pathway (UBE2S, USP32, MDM2, and UBE2C), genes encoding for kinases (MAP4K1, PIM1, and NEK2), and genes involved in RAS signaling (RANBP1 and RAB10).
Conclusions and next steps
“It seems that there’s some kind of crosstalk between the organ microenvironment and leukemic cells that make the leukemic cells have the phenotype of breast cancer,” Dr Cunningham said. “And it may well be that relapse sometimes results from the presence of an undiagnosed collection of these cells.”
Therefore, Dr Cunningham suggests performing scans in treatment-resistant leukemia patients. If a patient relapses, and particularly if lactic dehydrogenase levels are increased, a scan might be in order.
“If we can recognize these tumors and cut them out, the patient could be cured, because we’re successful at treating the bone marrow,” Dr Cunningham said. “We’ve had very good bone marrow drugs for 50 years.”
For her part, Dr Cunningham is delving further into this phenomenon. She is now conducting gene expression studies on the rest of the 25 leukemic breast tumor samples and comparing these tumors to breast cancer to identify the most significant dysregulated genes in both entities. The long-term goal is to find a way to predict which patients will develop leukemic breast tumors. ![]()
*Information in the abstract differs from that presented at the meeting.
SAN DIEGO—One woman’s curiosity and self-described “aggressive” approach to research have led to some unexpected discoveries about acute leukemias.
Isabel Cunningham, MD, of Columbia University in New York, has found evidence to suggest that treatment resistance in leukemia patients may sometimes result from an interaction between leukemic cells and the breast.
She discovered that leukemic cells in extramedullary niches can adopt a tumor phenotype similar to breast cancer.
And many genes are similarly upregulated in leukemic and epithelial breast tumors.
Her research indicates that a new approach to resistant leukemias that incorporates the principles of solid-tumor treatment—scans to identify any tumors and surgery to remove them—could decrease marrow relapse and death.
Dr Cunningham and her colleagues presented these findings in a poster at the AACR Annual Meeting 2014 (abstract 3996*).
“Chemotherapy resistance is our main problem in treating leukemia,” Dr Cunningham said. “It’s been known for a long time that, occasionally, leukemia forms tumors in an organ, but there’s never been a unified approach to treatment, except for leukemia that occurs in the testis and the meninges.”
Dr Cunningham had encountered many patients with resistant leukemia throughout her career, but her research actually began with a patient she had never met. A case study of a leukemia patient with a breast tumor sparked Dr Cunningham’s interest, and she emailed the study’s author to find out what ultimately became of the patient.
The response she received peaked her curiosity further. So she began seeking more of these cases, contacting authors, and collecting information on this phenomenon.
“I took this on as sort of a hobby,” Dr Cunningham said. “I never had any idea where this was going to lead.”
Eventually, she had amassed information on 235 cases—163 patients with acute myeloid leukemia (AML) and 72 with acute lymphoblastic leukemia (ALL)—who ranged from 1 year to 75 years of age. And an analysis of these cases led to some surprising discoveries.
Clinical findings
Dr Cunningham found these leukemic breast tumors can occur before, during, or after marrow leukemia. And, clinically, they resemble breast cancer. Most tumors were palpable, and some were detected only on routine mammograms.
There were single or multiple nodules that may have involved the entire breast. Sixty percent of cases were unilateral on presentation, but, often, the other breast became involved. Seventy percent of cases exhibited axillary lymphadenopathy that was ipsilateral.
Most tumors grew rapidly, to as large as 12 cm. The tumor behavior was similar in AML and ALL. And the tumors had a metastatic pattern similar to lobular breast cancer—spreading to the contralateral breast, the abdomen or pelvis, the meninges, and culminating in death.
However, some patients did survive. Four percent of patients who were treated only with chemotherapy were alive at 4 years. Twenty-five percent of patients had their tumors excised prior to chemotherapy and were alive anywhere from 3 years to more than 26 years after treatment.
Histology and gene expression
To build upon these findings, Dr Cunningham set her sights on patient samples. She was able to obtain paraffin blocks of leukemic breast tumors from 25 patients and perform immunohistochemical staining.
“It became clear that the leukemic tumors—which are marked by leukemic markers and not breast cancer markers—look, histologically, like breast cancer, specifically, lobular breast cancer,” Dr Cunningham said. “An additional pathologic finding was a specific type of desmoplastic fibrosis seen in all 25 contributed biopsies.”
Dr Cunningham also performed gene expression studies on 3 of the tumors (2 ALL and 1 AML), which were collected 8 months to 22 months after diagnosis, while marrows were in remission. The analyses revealed that a number of genes are significantly upregulated in both leukemic breast tumors and breast cancer.
These include genes involved in adhesion and interactions with the extracellular matrix (ADAM8, COMP, and CDH22), genes involved in the ubiquitin-proteasome pathway (UBE2S, USP32, MDM2, and UBE2C), genes encoding for kinases (MAP4K1, PIM1, and NEK2), and genes involved in RAS signaling (RANBP1 and RAB10).
Conclusions and next steps
“It seems that there’s some kind of crosstalk between the organ microenvironment and leukemic cells that make the leukemic cells have the phenotype of breast cancer,” Dr Cunningham said. “And it may well be that relapse sometimes results from the presence of an undiagnosed collection of these cells.”
Therefore, Dr Cunningham suggests performing scans in treatment-resistant leukemia patients. If a patient relapses, and particularly if lactic dehydrogenase levels are increased, a scan might be in order.
“If we can recognize these tumors and cut them out, the patient could be cured, because we’re successful at treating the bone marrow,” Dr Cunningham said. “We’ve had very good bone marrow drugs for 50 years.”
For her part, Dr Cunningham is delving further into this phenomenon. She is now conducting gene expression studies on the rest of the 25 leukemic breast tumor samples and comparing these tumors to breast cancer to identify the most significant dysregulated genes in both entities. The long-term goal is to find a way to predict which patients will develop leukemic breast tumors. ![]()
*Information in the abstract differs from that presented at the meeting.
SAN DIEGO—One woman’s curiosity and self-described “aggressive” approach to research have led to some unexpected discoveries about acute leukemias.
Isabel Cunningham, MD, of Columbia University in New York, has found evidence to suggest that treatment resistance in leukemia patients may sometimes result from an interaction between leukemic cells and the breast.
She discovered that leukemic cells in extramedullary niches can adopt a tumor phenotype similar to breast cancer.
And many genes are similarly upregulated in leukemic and epithelial breast tumors.
Her research indicates that a new approach to resistant leukemias that incorporates the principles of solid-tumor treatment—scans to identify any tumors and surgery to remove them—could decrease marrow relapse and death.
Dr Cunningham and her colleagues presented these findings in a poster at the AACR Annual Meeting 2014 (abstract 3996*).
“Chemotherapy resistance is our main problem in treating leukemia,” Dr Cunningham said. “It’s been known for a long time that, occasionally, leukemia forms tumors in an organ, but there’s never been a unified approach to treatment, except for leukemia that occurs in the testis and the meninges.”
Dr Cunningham had encountered many patients with resistant leukemia throughout her career, but her research actually began with a patient she had never met. A case study of a leukemia patient with a breast tumor sparked Dr Cunningham’s interest, and she emailed the study’s author to find out what ultimately became of the patient.
The response she received peaked her curiosity further. So she began seeking more of these cases, contacting authors, and collecting information on this phenomenon.
“I took this on as sort of a hobby,” Dr Cunningham said. “I never had any idea where this was going to lead.”
Eventually, she had amassed information on 235 cases—163 patients with acute myeloid leukemia (AML) and 72 with acute lymphoblastic leukemia (ALL)—who ranged from 1 year to 75 years of age. And an analysis of these cases led to some surprising discoveries.
Clinical findings
Dr Cunningham found these leukemic breast tumors can occur before, during, or after marrow leukemia. And, clinically, they resemble breast cancer. Most tumors were palpable, and some were detected only on routine mammograms.
There were single or multiple nodules that may have involved the entire breast. Sixty percent of cases were unilateral on presentation, but, often, the other breast became involved. Seventy percent of cases exhibited axillary lymphadenopathy that was ipsilateral.
Most tumors grew rapidly, to as large as 12 cm. The tumor behavior was similar in AML and ALL. And the tumors had a metastatic pattern similar to lobular breast cancer—spreading to the contralateral breast, the abdomen or pelvis, the meninges, and culminating in death.
However, some patients did survive. Four percent of patients who were treated only with chemotherapy were alive at 4 years. Twenty-five percent of patients had their tumors excised prior to chemotherapy and were alive anywhere from 3 years to more than 26 years after treatment.
Histology and gene expression
To build upon these findings, Dr Cunningham set her sights on patient samples. She was able to obtain paraffin blocks of leukemic breast tumors from 25 patients and perform immunohistochemical staining.
“It became clear that the leukemic tumors—which are marked by leukemic markers and not breast cancer markers—look, histologically, like breast cancer, specifically, lobular breast cancer,” Dr Cunningham said. “An additional pathologic finding was a specific type of desmoplastic fibrosis seen in all 25 contributed biopsies.”
Dr Cunningham also performed gene expression studies on 3 of the tumors (2 ALL and 1 AML), which were collected 8 months to 22 months after diagnosis, while marrows were in remission. The analyses revealed that a number of genes are significantly upregulated in both leukemic breast tumors and breast cancer.
These include genes involved in adhesion and interactions with the extracellular matrix (ADAM8, COMP, and CDH22), genes involved in the ubiquitin-proteasome pathway (UBE2S, USP32, MDM2, and UBE2C), genes encoding for kinases (MAP4K1, PIM1, and NEK2), and genes involved in RAS signaling (RANBP1 and RAB10).
Conclusions and next steps
“It seems that there’s some kind of crosstalk between the organ microenvironment and leukemic cells that make the leukemic cells have the phenotype of breast cancer,” Dr Cunningham said. “And it may well be that relapse sometimes results from the presence of an undiagnosed collection of these cells.”
Therefore, Dr Cunningham suggests performing scans in treatment-resistant leukemia patients. If a patient relapses, and particularly if lactic dehydrogenase levels are increased, a scan might be in order.
“If we can recognize these tumors and cut them out, the patient could be cured, because we’re successful at treating the bone marrow,” Dr Cunningham said. “We’ve had very good bone marrow drugs for 50 years.”
For her part, Dr Cunningham is delving further into this phenomenon. She is now conducting gene expression studies on the rest of the 25 leukemic breast tumor samples and comparing these tumors to breast cancer to identify the most significant dysregulated genes in both entities. The long-term goal is to find a way to predict which patients will develop leukemic breast tumors. ![]()
*Information in the abstract differs from that presented at the meeting.
FDA lifts clinical hold on CAR T-cell study

Credit: MSKCC
The US Food and Drug Administration (FDA) has lifted a clinical hold placed on a trial of chimeric antigen receptor (CAR) T-cell therapy, according to one of the study’s investigators.
The study is an evaluation of 19-28z CAR T cells in patients with B-cell acute lymphoblastic leukemia.
Trial enrollment was halted after 2 patients died from complications related to cytokine release syndrome, within 2 weeks of receiving CAR T-cell therapy.
Investigator Michel Sadelain, MD, PhD, of Memorial Sloan-Kettering Cancer Center (MSKCC) in New York, said these deaths have been reviewed, trial enrollment criteria have been changed, and the clinical hold has been lifted.
The FDA put the trial on hold last month, after researchers at MSKCC notified the agency of the deaths and had decided to halt trial enrollment themselves.
Though several other patients have died on this study, only 2 of the deaths had unexpected causes and prompted additional investigation. One of these patients died of cardiovascular disease, and the other died following “persistent seizure activity.”
So MSKCC conducted a review of these cases. And the results prompted them to amend trial enrollment criteria and dosing recommendations. Now, patients with cardiac disease are ineligible to receive 19-28z CAR T cells.
And the T-cell dose a patient receives will depend on the extent of his or her disease. The hope is that this will reduce the risk of cytokine release syndrome and any resulting seizures.
The researchers also noted that the monoclonal antibody tocilizumab has proven effective in treating cytokine release syndrome. ![]()

Credit: MSKCC
The US Food and Drug Administration (FDA) has lifted a clinical hold placed on a trial of chimeric antigen receptor (CAR) T-cell therapy, according to one of the study’s investigators.
The study is an evaluation of 19-28z CAR T cells in patients with B-cell acute lymphoblastic leukemia.
Trial enrollment was halted after 2 patients died from complications related to cytokine release syndrome, within 2 weeks of receiving CAR T-cell therapy.
Investigator Michel Sadelain, MD, PhD, of Memorial Sloan-Kettering Cancer Center (MSKCC) in New York, said these deaths have been reviewed, trial enrollment criteria have been changed, and the clinical hold has been lifted.
The FDA put the trial on hold last month, after researchers at MSKCC notified the agency of the deaths and had decided to halt trial enrollment themselves.
Though several other patients have died on this study, only 2 of the deaths had unexpected causes and prompted additional investigation. One of these patients died of cardiovascular disease, and the other died following “persistent seizure activity.”
So MSKCC conducted a review of these cases. And the results prompted them to amend trial enrollment criteria and dosing recommendations. Now, patients with cardiac disease are ineligible to receive 19-28z CAR T cells.
And the T-cell dose a patient receives will depend on the extent of his or her disease. The hope is that this will reduce the risk of cytokine release syndrome and any resulting seizures.
The researchers also noted that the monoclonal antibody tocilizumab has proven effective in treating cytokine release syndrome. ![]()

Credit: MSKCC
The US Food and Drug Administration (FDA) has lifted a clinical hold placed on a trial of chimeric antigen receptor (CAR) T-cell therapy, according to one of the study’s investigators.
The study is an evaluation of 19-28z CAR T cells in patients with B-cell acute lymphoblastic leukemia.
Trial enrollment was halted after 2 patients died from complications related to cytokine release syndrome, within 2 weeks of receiving CAR T-cell therapy.
Investigator Michel Sadelain, MD, PhD, of Memorial Sloan-Kettering Cancer Center (MSKCC) in New York, said these deaths have been reviewed, trial enrollment criteria have been changed, and the clinical hold has been lifted.
The FDA put the trial on hold last month, after researchers at MSKCC notified the agency of the deaths and had decided to halt trial enrollment themselves.
Though several other patients have died on this study, only 2 of the deaths had unexpected causes and prompted additional investigation. One of these patients died of cardiovascular disease, and the other died following “persistent seizure activity.”
So MSKCC conducted a review of these cases. And the results prompted them to amend trial enrollment criteria and dosing recommendations. Now, patients with cardiac disease are ineligible to receive 19-28z CAR T cells.
And the T-cell dose a patient receives will depend on the extent of his or her disease. The hope is that this will reduce the risk of cytokine release syndrome and any resulting seizures.
The researchers also noted that the monoclonal antibody tocilizumab has proven effective in treating cytokine release syndrome. ![]()
Explaining the link between Down syndrome and B-ALL
Credit: Aaron Logan
Investigators believe they’ve uncovered information that explains the connection between Down syndrome and B-cell acute lymphoblastic leukemia (B-ALL).
In a letter to Nature Genetics, the team described how they tracked the chain of events that links a chromosomal abnormality in Down syndrome to the cellular havoc that occurs in B-cell ALL.
Experiments in mice and patient samples revealed that the gene HMGN1 turns off the function of PRC2, which prompts B-cell proliferation.
“For 80 years, it hasn’t been clear why children with Down syndrome face a sharply elevated risk of ALL,” said the study’s lead author, Andrew Lane, MD, PhD, of Dana-Farber Cancer Institute in Boston.
“Advances in technology—which make it possible to study blood cells and leukemias that model Down syndrome in the laboratory—have enabled us to make that link.”
To trace the link between Down syndrome and B-ALL, the investigators acquired a strain of mice that carry an extra copy of 31 genes found on chromosome 21. B cells from these mice were abnormal and grew uncontrollably, just as they do in B-ALL patients.
The team set out to characterize the pattern of gene activity that distinguishes these abnormal B cells from normal B cells. They found the chief difference was that, in the abnormal cells, PRC2 proteins did not function. Somehow, the loss of PRC2 was spurring the B cells to divide and proliferate before they were fully mature.
To confirm that a shutdown of PRC2 is critical to the formation of B-ALL in Down syndrome patients, the investigators focused on the genes controlled by PRC2. Using 2 sets of B-ALL cell samples—1 from patients with Down syndrome and 1 from patients without it—they measured the activity of thousands of different genes, looking for differences between the 2 sets.
About 100 genes were much more active in the Down syndrome group, and all of them were under the control of PRC2. When PRC2 is silenced, those 100 genes respond with a burst of activity, driving cell growth and division.
The investigators then wondered what gene or group of genes was stifling PRC2 in Down syndrome patients’ B cells. Using cells from the mouse models, the team systematically switched off each of the 31 genes to determine its effect on the cells. When they turned off HMGN1, the cells stopped growing and died.
“We concluded that the extra copy of HMGN1 is important for turning off PRC2, and that, in turn, increases the cell proliferation,” Dr Lane said. “This provides the long-sought-after molecular link between Down syndrome and the development of B-cell ALL.”
Although there are currently no drugs that target HMGN1, the investigators suggest HDAC inhibitors that switch on PRC2 could have an antileukemic effect in patients with Down syndrome. Work is under way to improve these drugs so they can be tested in preclinical experiments.
As other forms of B-ALL also have the same 100-gene signature as the one discovered for B-ALL associated with Down syndrome, agents that target PRC2 might be effective in those cancers as well, Dr Lane noted. ![]()

Credit: Aaron Logan
Investigators believe they’ve uncovered information that explains the connection between Down syndrome and B-cell acute lymphoblastic leukemia (B-ALL).
In a letter to Nature Genetics, the team described how they tracked the chain of events that links a chromosomal abnormality in Down syndrome to the cellular havoc that occurs in B-cell ALL.
Experiments in mice and patient samples revealed that the gene HMGN1 turns off the function of PRC2, which prompts B-cell proliferation.
“For 80 years, it hasn’t been clear why children with Down syndrome face a sharply elevated risk of ALL,” said the study’s lead author, Andrew Lane, MD, PhD, of Dana-Farber Cancer Institute in Boston.
“Advances in technology—which make it possible to study blood cells and leukemias that model Down syndrome in the laboratory—have enabled us to make that link.”
To trace the link between Down syndrome and B-ALL, the investigators acquired a strain of mice that carry an extra copy of 31 genes found on chromosome 21. B cells from these mice were abnormal and grew uncontrollably, just as they do in B-ALL patients.
The team set out to characterize the pattern of gene activity that distinguishes these abnormal B cells from normal B cells. They found the chief difference was that, in the abnormal cells, PRC2 proteins did not function. Somehow, the loss of PRC2 was spurring the B cells to divide and proliferate before they were fully mature.
To confirm that a shutdown of PRC2 is critical to the formation of B-ALL in Down syndrome patients, the investigators focused on the genes controlled by PRC2. Using 2 sets of B-ALL cell samples—1 from patients with Down syndrome and 1 from patients without it—they measured the activity of thousands of different genes, looking for differences between the 2 sets.
About 100 genes were much more active in the Down syndrome group, and all of them were under the control of PRC2. When PRC2 is silenced, those 100 genes respond with a burst of activity, driving cell growth and division.
The investigators then wondered what gene or group of genes was stifling PRC2 in Down syndrome patients’ B cells. Using cells from the mouse models, the team systematically switched off each of the 31 genes to determine its effect on the cells. When they turned off HMGN1, the cells stopped growing and died.
“We concluded that the extra copy of HMGN1 is important for turning off PRC2, and that, in turn, increases the cell proliferation,” Dr Lane said. “This provides the long-sought-after molecular link between Down syndrome and the development of B-cell ALL.”
Although there are currently no drugs that target HMGN1, the investigators suggest HDAC inhibitors that switch on PRC2 could have an antileukemic effect in patients with Down syndrome. Work is under way to improve these drugs so they can be tested in preclinical experiments.
As other forms of B-ALL also have the same 100-gene signature as the one discovered for B-ALL associated with Down syndrome, agents that target PRC2 might be effective in those cancers as well, Dr Lane noted. ![]()

Credit: Aaron Logan
Investigators believe they’ve uncovered information that explains the connection between Down syndrome and B-cell acute lymphoblastic leukemia (B-ALL).
In a letter to Nature Genetics, the team described how they tracked the chain of events that links a chromosomal abnormality in Down syndrome to the cellular havoc that occurs in B-cell ALL.
Experiments in mice and patient samples revealed that the gene HMGN1 turns off the function of PRC2, which prompts B-cell proliferation.
“For 80 years, it hasn’t been clear why children with Down syndrome face a sharply elevated risk of ALL,” said the study’s lead author, Andrew Lane, MD, PhD, of Dana-Farber Cancer Institute in Boston.
“Advances in technology—which make it possible to study blood cells and leukemias that model Down syndrome in the laboratory—have enabled us to make that link.”
To trace the link between Down syndrome and B-ALL, the investigators acquired a strain of mice that carry an extra copy of 31 genes found on chromosome 21. B cells from these mice were abnormal and grew uncontrollably, just as they do in B-ALL patients.
The team set out to characterize the pattern of gene activity that distinguishes these abnormal B cells from normal B cells. They found the chief difference was that, in the abnormal cells, PRC2 proteins did not function. Somehow, the loss of PRC2 was spurring the B cells to divide and proliferate before they were fully mature.
To confirm that a shutdown of PRC2 is critical to the formation of B-ALL in Down syndrome patients, the investigators focused on the genes controlled by PRC2. Using 2 sets of B-ALL cell samples—1 from patients with Down syndrome and 1 from patients without it—they measured the activity of thousands of different genes, looking for differences between the 2 sets.
About 100 genes were much more active in the Down syndrome group, and all of them were under the control of PRC2. When PRC2 is silenced, those 100 genes respond with a burst of activity, driving cell growth and division.
The investigators then wondered what gene or group of genes was stifling PRC2 in Down syndrome patients’ B cells. Using cells from the mouse models, the team systematically switched off each of the 31 genes to determine its effect on the cells. When they turned off HMGN1, the cells stopped growing and died.
“We concluded that the extra copy of HMGN1 is important for turning off PRC2, and that, in turn, increases the cell proliferation,” Dr Lane said. “This provides the long-sought-after molecular link between Down syndrome and the development of B-cell ALL.”
Although there are currently no drugs that target HMGN1, the investigators suggest HDAC inhibitors that switch on PRC2 could have an antileukemic effect in patients with Down syndrome. Work is under way to improve these drugs so they can be tested in preclinical experiments.
As other forms of B-ALL also have the same 100-gene signature as the one discovered for B-ALL associated with Down syndrome, agents that target PRC2 might be effective in those cancers as well, Dr Lane noted. ![]()
Enrollment stalled for CAR T-cell study
Update: The hold on this trial has been lifted. Click here for additional details.
Memorial Sloan-Kettering Cancer Center has temporarily suspended enrollment in a study of chimeric antigen receptor (CAR) T-cell therapy, due to 2 patient deaths.
The study is an evaluation of CD19-targeted CAR T cells in patients with B-cell acute lymphoblastic leukemia (ALL).
Of the 22 patients enrolled on the study to date, 10 have died. But only 2 of these deaths gave researchers pause and made them question enrollment criteria.
Six deaths were a result of disease relapse or progression, and 2 patients died of complications from stem cell transplant.
The 2 deaths that prompted the suspension of enrollment occurred within 2 weeks of the patients receiving CAR T cells.
“The first of these patients had a prior history of cardiac disease, while the second patient died due to complications associated with persistent seizure activity,” said Renier Brentjens, MD, PhD, of Memorial Sloan-Kettering in New York.
“As a matter of routine review at Sloan-Kettering for adverse events on-study, our center made the decision to pause enrollment and review these 2 patients in greater detail.”
“And as a consequence of this review, we’ve amended the enrollment criteria in regards to comorbidities, thereby excluding patients with cardiac disease, and adjusted the T-cell dose based on the extent of disease, [in the] hope that this modification will reduce the cytokine release syndrome that these patients with morphological disease have experienced.”
The researchers expect the trial to resume enrollment soon.
Some results from this study were recently published in Science Translational Medicine, and Dr Brentjens presented the latest results at the AACR Annual Meeting 2014 in San Diego (abstract CT102*).
Thus far, the researchers have enrolled 22 adult patients who had relapsed or refractory B-ALL, were minimal residual disease-positive, or were in the first complete remission (CR1) at enrollment. Patients in CR1 were monitored and only received CAR T cells if they relapsed.
The remaining patients received re-induction chemotherapy (physician’s choice), followed by CAR T-cell infusion. After treatment, the options were allogeneic transplant, a different salvage therapy, or monitoring.
In all, 20 patients received a CAR T-cell dose of 3 x 106 T cells/kg. Eighty-two percent of patients initially achieved a CR, and 72% had a morphologic CR. The average time to CR was about 24.5 days.
Twelve of the responders were eligible for transplant. Of the 8 patients who ultimately underwent transplant and survived, 1 relapsed, but the rest remain in remission.
Dr Brentjens noted that some patients developed cytokine release syndrome, and this was related to the amount of disease present at the time of CAR T-cell infusion.
“Those patients that had only minimal residual disease at the time of CAR T-cell infusion . . . less than 5% blasts, generally had either no fever or very transient, low-grade fever,” he said.
“In contrast, all those patients that had morphologic residual disease at the time of CAR T-cell infusion demonstrated a high, persistent spike in fevers . . . , became hypotensive, and required transfer—for additional, closer monitoring—to our ICU.”
The researchers initially treated these patients with high-dose steroids, which reduced cytokine levels in the serum and ameliorated fevers. But it also rapidly reduced T-cell populations to undetectable levels.
Fortunately, another group of researchers subsequently discovered that the monoclonal antibody tocilizumab can treat cytokine release syndrome without inducing this side effect. So Dr Brentjens and his colleagues began using this drug and found it both safe and effective. ![]()
*Information in the abstract differs from that presented at the meeting.
Update: The hold on this trial has been lifted. Click here for additional details.
Memorial Sloan-Kettering Cancer Center has temporarily suspended enrollment in a study of chimeric antigen receptor (CAR) T-cell therapy, due to 2 patient deaths.
The study is an evaluation of CD19-targeted CAR T cells in patients with B-cell acute lymphoblastic leukemia (ALL).
Of the 22 patients enrolled on the study to date, 10 have died. But only 2 of these deaths gave researchers pause and made them question enrollment criteria.
Six deaths were a result of disease relapse or progression, and 2 patients died of complications from stem cell transplant.
The 2 deaths that prompted the suspension of enrollment occurred within 2 weeks of the patients receiving CAR T cells.
“The first of these patients had a prior history of cardiac disease, while the second patient died due to complications associated with persistent seizure activity,” said Renier Brentjens, MD, PhD, of Memorial Sloan-Kettering in New York.
“As a matter of routine review at Sloan-Kettering for adverse events on-study, our center made the decision to pause enrollment and review these 2 patients in greater detail.”
“And as a consequence of this review, we’ve amended the enrollment criteria in regards to comorbidities, thereby excluding patients with cardiac disease, and adjusted the T-cell dose based on the extent of disease, [in the] hope that this modification will reduce the cytokine release syndrome that these patients with morphological disease have experienced.”
The researchers expect the trial to resume enrollment soon.
Some results from this study were recently published in Science Translational Medicine, and Dr Brentjens presented the latest results at the AACR Annual Meeting 2014 in San Diego (abstract CT102*).
Thus far, the researchers have enrolled 22 adult patients who had relapsed or refractory B-ALL, were minimal residual disease-positive, or were in the first complete remission (CR1) at enrollment. Patients in CR1 were monitored and only received CAR T cells if they relapsed.
The remaining patients received re-induction chemotherapy (physician’s choice), followed by CAR T-cell infusion. After treatment, the options were allogeneic transplant, a different salvage therapy, or monitoring.
In all, 20 patients received a CAR T-cell dose of 3 x 106 T cells/kg. Eighty-two percent of patients initially achieved a CR, and 72% had a morphologic CR. The average time to CR was about 24.5 days.
Twelve of the responders were eligible for transplant. Of the 8 patients who ultimately underwent transplant and survived, 1 relapsed, but the rest remain in remission.
Dr Brentjens noted that some patients developed cytokine release syndrome, and this was related to the amount of disease present at the time of CAR T-cell infusion.
“Those patients that had only minimal residual disease at the time of CAR T-cell infusion . . . less than 5% blasts, generally had either no fever or very transient, low-grade fever,” he said.
“In contrast, all those patients that had morphologic residual disease at the time of CAR T-cell infusion demonstrated a high, persistent spike in fevers . . . , became hypotensive, and required transfer—for additional, closer monitoring—to our ICU.”
The researchers initially treated these patients with high-dose steroids, which reduced cytokine levels in the serum and ameliorated fevers. But it also rapidly reduced T-cell populations to undetectable levels.
Fortunately, another group of researchers subsequently discovered that the monoclonal antibody tocilizumab can treat cytokine release syndrome without inducing this side effect. So Dr Brentjens and his colleagues began using this drug and found it both safe and effective. ![]()
*Information in the abstract differs from that presented at the meeting.
Update: The hold on this trial has been lifted. Click here for additional details.
Memorial Sloan-Kettering Cancer Center has temporarily suspended enrollment in a study of chimeric antigen receptor (CAR) T-cell therapy, due to 2 patient deaths.
The study is an evaluation of CD19-targeted CAR T cells in patients with B-cell acute lymphoblastic leukemia (ALL).
Of the 22 patients enrolled on the study to date, 10 have died. But only 2 of these deaths gave researchers pause and made them question enrollment criteria.
Six deaths were a result of disease relapse or progression, and 2 patients died of complications from stem cell transplant.
The 2 deaths that prompted the suspension of enrollment occurred within 2 weeks of the patients receiving CAR T cells.
“The first of these patients had a prior history of cardiac disease, while the second patient died due to complications associated with persistent seizure activity,” said Renier Brentjens, MD, PhD, of Memorial Sloan-Kettering in New York.
“As a matter of routine review at Sloan-Kettering for adverse events on-study, our center made the decision to pause enrollment and review these 2 patients in greater detail.”
“And as a consequence of this review, we’ve amended the enrollment criteria in regards to comorbidities, thereby excluding patients with cardiac disease, and adjusted the T-cell dose based on the extent of disease, [in the] hope that this modification will reduce the cytokine release syndrome that these patients with morphological disease have experienced.”
The researchers expect the trial to resume enrollment soon.
Some results from this study were recently published in Science Translational Medicine, and Dr Brentjens presented the latest results at the AACR Annual Meeting 2014 in San Diego (abstract CT102*).
Thus far, the researchers have enrolled 22 adult patients who had relapsed or refractory B-ALL, were minimal residual disease-positive, or were in the first complete remission (CR1) at enrollment. Patients in CR1 were monitored and only received CAR T cells if they relapsed.
The remaining patients received re-induction chemotherapy (physician’s choice), followed by CAR T-cell infusion. After treatment, the options were allogeneic transplant, a different salvage therapy, or monitoring.
In all, 20 patients received a CAR T-cell dose of 3 x 106 T cells/kg. Eighty-two percent of patients initially achieved a CR, and 72% had a morphologic CR. The average time to CR was about 24.5 days.
Twelve of the responders were eligible for transplant. Of the 8 patients who ultimately underwent transplant and survived, 1 relapsed, but the rest remain in remission.
Dr Brentjens noted that some patients developed cytokine release syndrome, and this was related to the amount of disease present at the time of CAR T-cell infusion.
“Those patients that had only minimal residual disease at the time of CAR T-cell infusion . . . less than 5% blasts, generally had either no fever or very transient, low-grade fever,” he said.
“In contrast, all those patients that had morphologic residual disease at the time of CAR T-cell infusion demonstrated a high, persistent spike in fevers . . . , became hypotensive, and required transfer—for additional, closer monitoring—to our ICU.”
The researchers initially treated these patients with high-dose steroids, which reduced cytokine levels in the serum and ameliorated fevers. But it also rapidly reduced T-cell populations to undetectable levels.
Fortunately, another group of researchers subsequently discovered that the monoclonal antibody tocilizumab can treat cytokine release syndrome without inducing this side effect. So Dr Brentjens and his colleagues began using this drug and found it both safe and effective. ![]()
*Information in the abstract differs from that presented at the meeting.
ALL cells don’t survive SSC culture method
Results of a pilot study suggest safe transplantation of spermatogonial stem cells (SSCs) may be possible, potentially bringing us one step closer to ensuring fertility preservation in young boys with cancer.
Investigators found they could culture a large amount of SSCs using testicular tissue from boys with acute lymphoblastic leukemia (ALL).
And the ALL cells did not survive the culture process.
The researchers reported these results in Fertility and Sterility.
“Our study addressed an important safety issue—whether cancer cells that might be present in testicular tissue samples can survive the process to replicate the sperm-producing stem cells,” said study author Hooman Sadri-Ardekani, MD, PhD, of Wake Forest Baptist Medical Center in Winston-Salem, North Carolina.
“This is an important consideration because of the potential to re-introduce cancer into the patient. The research, which involved one of the most common childhood cancers, shows that the cancer cells were eliminated. Based on these findings, we recommend that all boys with cancer be offered the option of storing testicular tissue for possible future clinical use.”
Previous research had shown that up to 30% of boys with ALL had cancer cells in their testicular tissue. In several studies, researchers attempted to eliminate cancer cells from biopsy tissue, but the results varied.
So Dr Sadri-Ardekani and his colleagues decided to investigate whether cancer cells would survive the protocol they had developed to reproduce SSCs from a small tissue biopsy.
This process multiplies the original SSCs by 18,000-fold so there are enough cells to transplant back into the patient when he reaches adulthood.
The investigators tested the method using samples from 3 ALL patients. The team cultured the ALL cells alone and at various concentrations in combination with testicular cells.
The ALL cells cultured alone did not survive beyond 14 days. But the testicular cells cultured in parallel survived and continue to proliferate well after 8 weeks.
At 10 to 16 days, the ALL cells cultured in combination had begun to die off. The cells were undetectable in cultures from 2 of the patients that had an initial ALL concentration of 0.04%, 0.4%, or 4%.
At 20 to 26 days, ALL cells were undetectable in all cultures, even those with an initial concentration of 40% ALL cells.
“This pilot study showed that the culture system not only allowed for efficient propagation of sperm stem cells but also eliminated ALL cells,” Dr Sadri-Ardekani said.
SSC transplants have not yet been attempted in humans, but they have been performed successfully in several species of animals, including monkeys.
Dr Sadri-Ardekani noted that before we can perform SSC transplants in patients, additional research is needed, particularly, research investigating whether other types of leukemia cells will also be eliminated in the cell-propagation process. ![]()

Results of a pilot study suggest safe transplantation of spermatogonial stem cells (SSCs) may be possible, potentially bringing us one step closer to ensuring fertility preservation in young boys with cancer.
Investigators found they could culture a large amount of SSCs using testicular tissue from boys with acute lymphoblastic leukemia (ALL).
And the ALL cells did not survive the culture process.
The researchers reported these results in Fertility and Sterility.
“Our study addressed an important safety issue—whether cancer cells that might be present in testicular tissue samples can survive the process to replicate the sperm-producing stem cells,” said study author Hooman Sadri-Ardekani, MD, PhD, of Wake Forest Baptist Medical Center in Winston-Salem, North Carolina.
“This is an important consideration because of the potential to re-introduce cancer into the patient. The research, which involved one of the most common childhood cancers, shows that the cancer cells were eliminated. Based on these findings, we recommend that all boys with cancer be offered the option of storing testicular tissue for possible future clinical use.”
Previous research had shown that up to 30% of boys with ALL had cancer cells in their testicular tissue. In several studies, researchers attempted to eliminate cancer cells from biopsy tissue, but the results varied.
So Dr Sadri-Ardekani and his colleagues decided to investigate whether cancer cells would survive the protocol they had developed to reproduce SSCs from a small tissue biopsy.
This process multiplies the original SSCs by 18,000-fold so there are enough cells to transplant back into the patient when he reaches adulthood.
The investigators tested the method using samples from 3 ALL patients. The team cultured the ALL cells alone and at various concentrations in combination with testicular cells.
The ALL cells cultured alone did not survive beyond 14 days. But the testicular cells cultured in parallel survived and continue to proliferate well after 8 weeks.
At 10 to 16 days, the ALL cells cultured in combination had begun to die off. The cells were undetectable in cultures from 2 of the patients that had an initial ALL concentration of 0.04%, 0.4%, or 4%.
At 20 to 26 days, ALL cells were undetectable in all cultures, even those with an initial concentration of 40% ALL cells.
“This pilot study showed that the culture system not only allowed for efficient propagation of sperm stem cells but also eliminated ALL cells,” Dr Sadri-Ardekani said.
SSC transplants have not yet been attempted in humans, but they have been performed successfully in several species of animals, including monkeys.
Dr Sadri-Ardekani noted that before we can perform SSC transplants in patients, additional research is needed, particularly, research investigating whether other types of leukemia cells will also be eliminated in the cell-propagation process. ![]()

Results of a pilot study suggest safe transplantation of spermatogonial stem cells (SSCs) may be possible, potentially bringing us one step closer to ensuring fertility preservation in young boys with cancer.
Investigators found they could culture a large amount of SSCs using testicular tissue from boys with acute lymphoblastic leukemia (ALL).
And the ALL cells did not survive the culture process.
The researchers reported these results in Fertility and Sterility.
“Our study addressed an important safety issue—whether cancer cells that might be present in testicular tissue samples can survive the process to replicate the sperm-producing stem cells,” said study author Hooman Sadri-Ardekani, MD, PhD, of Wake Forest Baptist Medical Center in Winston-Salem, North Carolina.
“This is an important consideration because of the potential to re-introduce cancer into the patient. The research, which involved one of the most common childhood cancers, shows that the cancer cells were eliminated. Based on these findings, we recommend that all boys with cancer be offered the option of storing testicular tissue for possible future clinical use.”
Previous research had shown that up to 30% of boys with ALL had cancer cells in their testicular tissue. In several studies, researchers attempted to eliminate cancer cells from biopsy tissue, but the results varied.
So Dr Sadri-Ardekani and his colleagues decided to investigate whether cancer cells would survive the protocol they had developed to reproduce SSCs from a small tissue biopsy.
This process multiplies the original SSCs by 18,000-fold so there are enough cells to transplant back into the patient when he reaches adulthood.
The investigators tested the method using samples from 3 ALL patients. The team cultured the ALL cells alone and at various concentrations in combination with testicular cells.
The ALL cells cultured alone did not survive beyond 14 days. But the testicular cells cultured in parallel survived and continue to proliferate well after 8 weeks.
At 10 to 16 days, the ALL cells cultured in combination had begun to die off. The cells were undetectable in cultures from 2 of the patients that had an initial ALL concentration of 0.04%, 0.4%, or 4%.
At 20 to 26 days, ALL cells were undetectable in all cultures, even those with an initial concentration of 40% ALL cells.
“This pilot study showed that the culture system not only allowed for efficient propagation of sperm stem cells but also eliminated ALL cells,” Dr Sadri-Ardekani said.
SSC transplants have not yet been attempted in humans, but they have been performed successfully in several species of animals, including monkeys.
Dr Sadri-Ardekani noted that before we can perform SSC transplants in patients, additional research is needed, particularly, research investigating whether other types of leukemia cells will also be eliminated in the cell-propagation process. ![]()
Abnormality increases risk of ALL subtype
Credit: Beth A. Sullivan
Individuals with a rare chromosomal abnormality have a 2700-fold increased risk of developing a type of acute lymphoblastic leukemia (ALL), according to a study published in Nature.
Previous research showed that a small subset of ALL patients have recurrent amplification of megabase regions of chromosome 21.
In the current study, investigators decided to reconstruct the sequence of genetic events that lead to this type of leukemia, iAMP21 ALL.
They found that some patients with iAMP21 ALL had an abnormality in which chromosome 15 and chromosome 21 are fused together, known as a Robertsonian translocation.
And the joining of these 2 chromosomes increased a person’s risk of developing iAMP21 ALL 2700-fold, when compared to the general population.
“Although rare, people who carry this specific joining together of chromosomes 15 and 21 are specifically and massively predisposed to iAMP21 ALL,” said study author Christine Harrison, PhD, of Newcastle University in the UK.
“We have been able to map the roads the cells follow in their transition from a normal genome to a leukemia genome.”
To do this, the investigators sequenced 9 samples from patients with iAMP21 ALL, 4 with the rare Robertsonian translocation and 5 without it.
For the 4 patients with the translocation, their leukemia was initiated by chromothripsis. This event shatters a chromosome—in this case, the joined chromosomes 15 and 21—and then the DNA repair machinery pastes the chromosome back together in a highly flawed and inaccurate order.
In the 5 other patients, the cancer was initiated by 2 copies of chromosome 21 being fused together, head-to-head, usually followed by chromothripsis.
The investigators found a consistent sequence of genetic events across the patients studied. Although the events seem random and chaotic at first, the end result is a new chromosome 21 in which the numbers and arrangement of genes are optimized to drive leukemia.
“What is striking about our findings is that this type of leukemia could develop incredibly quickly—potentially in just a few rounds of cell division,” said study author Peter Campbell, PhD, of the Wellcome Trust Sanger Institute in Hinxton, UK.
“We now want to understand why the abnormally fused chromosomes are so susceptible to this catastrophic shattering.” ![]()

Credit: Beth A. Sullivan
Individuals with a rare chromosomal abnormality have a 2700-fold increased risk of developing a type of acute lymphoblastic leukemia (ALL), according to a study published in Nature.
Previous research showed that a small subset of ALL patients have recurrent amplification of megabase regions of chromosome 21.
In the current study, investigators decided to reconstruct the sequence of genetic events that lead to this type of leukemia, iAMP21 ALL.
They found that some patients with iAMP21 ALL had an abnormality in which chromosome 15 and chromosome 21 are fused together, known as a Robertsonian translocation.
And the joining of these 2 chromosomes increased a person’s risk of developing iAMP21 ALL 2700-fold, when compared to the general population.
“Although rare, people who carry this specific joining together of chromosomes 15 and 21 are specifically and massively predisposed to iAMP21 ALL,” said study author Christine Harrison, PhD, of Newcastle University in the UK.
“We have been able to map the roads the cells follow in their transition from a normal genome to a leukemia genome.”
To do this, the investigators sequenced 9 samples from patients with iAMP21 ALL, 4 with the rare Robertsonian translocation and 5 without it.
For the 4 patients with the translocation, their leukemia was initiated by chromothripsis. This event shatters a chromosome—in this case, the joined chromosomes 15 and 21—and then the DNA repair machinery pastes the chromosome back together in a highly flawed and inaccurate order.
In the 5 other patients, the cancer was initiated by 2 copies of chromosome 21 being fused together, head-to-head, usually followed by chromothripsis.
The investigators found a consistent sequence of genetic events across the patients studied. Although the events seem random and chaotic at first, the end result is a new chromosome 21 in which the numbers and arrangement of genes are optimized to drive leukemia.
“What is striking about our findings is that this type of leukemia could develop incredibly quickly—potentially in just a few rounds of cell division,” said study author Peter Campbell, PhD, of the Wellcome Trust Sanger Institute in Hinxton, UK.
“We now want to understand why the abnormally fused chromosomes are so susceptible to this catastrophic shattering.” ![]()

Credit: Beth A. Sullivan
Individuals with a rare chromosomal abnormality have a 2700-fold increased risk of developing a type of acute lymphoblastic leukemia (ALL), according to a study published in Nature.
Previous research showed that a small subset of ALL patients have recurrent amplification of megabase regions of chromosome 21.
In the current study, investigators decided to reconstruct the sequence of genetic events that lead to this type of leukemia, iAMP21 ALL.
They found that some patients with iAMP21 ALL had an abnormality in which chromosome 15 and chromosome 21 are fused together, known as a Robertsonian translocation.
And the joining of these 2 chromosomes increased a person’s risk of developing iAMP21 ALL 2700-fold, when compared to the general population.
“Although rare, people who carry this specific joining together of chromosomes 15 and 21 are specifically and massively predisposed to iAMP21 ALL,” said study author Christine Harrison, PhD, of Newcastle University in the UK.
“We have been able to map the roads the cells follow in their transition from a normal genome to a leukemia genome.”
To do this, the investigators sequenced 9 samples from patients with iAMP21 ALL, 4 with the rare Robertsonian translocation and 5 without it.
For the 4 patients with the translocation, their leukemia was initiated by chromothripsis. This event shatters a chromosome—in this case, the joined chromosomes 15 and 21—and then the DNA repair machinery pastes the chromosome back together in a highly flawed and inaccurate order.
In the 5 other patients, the cancer was initiated by 2 copies of chromosome 21 being fused together, head-to-head, usually followed by chromothripsis.
The investigators found a consistent sequence of genetic events across the patients studied. Although the events seem random and chaotic at first, the end result is a new chromosome 21 in which the numbers and arrangement of genes are optimized to drive leukemia.
“What is striking about our findings is that this type of leukemia could develop incredibly quickly—potentially in just a few rounds of cell division,” said study author Peter Campbell, PhD, of the Wellcome Trust Sanger Institute in Hinxton, UK.
“We now want to understand why the abnormally fused chromosomes are so susceptible to this catastrophic shattering.”
ALL maintenance phase still stressful, studies show

Credit: Bill Branson
The stress and lifestyle changes mothers experience after a child’s leukemia diagnosis do not disappear once the disease is in the maintenance phase, new research suggests.
One study showed that mothers of patients with acute lymphoblastic leukemia (ALL) continued to exhibit signs of stress, anxiety, and depression when their child’s disease was in the maintenance phase.
And another study showed that a family’s daily schedule and sleeping arrangements did not return to the way they were before the child’s diagnosis.
“Even though these mothers were in the maintenance phase of their child’s illness, and the prognosis was good, we heard them say over and over that things could never go back to what they were before,” said study author Madalynn Neu, PhD, RN, of the Colorado University College of Nursing.
Stress, depression, and anxiety
In the Journal of Pediatric Oncology Nursing, Dr Neu and her colleagues reported their analysis of stress, anxiety, and depression among the mothers of patients with ALL. The team evaluated 26 mothers of ALL patients and 26 mothers of healthy children, who were matched according to the child’s age and gender.
To assess anxiety, the researchers collected salivary cortisol from the mothers 4 times a day for 3 consecutive days. The mothers also completed questionnaires—the Hospital Anxiety and Depression Scale and the Perceived Stress Scale.
More mothers in the ALL group had questionnaire scores indicating clinical anxiety (46%) and depressive symptoms (27%). And there was a trend toward increased stress in the mothers of ALL patients.
However, the researchers were surprised to find that the mothers’ anxiety levels—as measured by salivary cortisol—were similar to the mothers of well children.
“This may have been affected by the fact that even the control group wasn’t without anxiety,” said study author Ellen Matthews, PhD, RN, of the Colorado University College of Nursing.
“Financial, marital, social, and career concerns mean that parents of young children experience anxiety even without ALL.”
The sleep-wake experience
In the Journal of Pediatric Nursing, the researchers described their assessment of the sleep-wake experience for mothers of ALL patients. The team conducted interviews with 20 mothers, using open-ended, semi-structured questions, and the answers were transcribed verbatim.
Two main themes emerged during these interviews. The first was dubbed, “It’s a whole new cancer world” and contained 4 subthemes: losing normality, being off-balance/insecure, juggling duties, and making transitions.
“Many [of the mothers] had lost their normal lives—lost jobs, houses, friends,” Dr Matthews said. “Some were juggling their time around their child’s needs, and they had fears about many things—fear of recurrence, fear of making a mistake with medication, fear their kids might get sick with an infection.”
The second theme that emerged in interviews was, “I don’t remember what it’s like to have sleep.” This also contained 4 subthemes: sleeping trouble before and after ALL, the child feeling sick at night, worrying, and coping with exhaustion.
The mothers also noted that once sleep arrangements had changed, they often did not return to their pretreatment “normal.”
“Mothers talked about the difficulty of sleep while giving steroid medication,” Dr Neu said. “And if the ill child got to stay up late watching movies, the siblings wanted to stay up too.”
“The same was true of sleeping in a parent’s room. If an ill child wanted to sleep close to a parent (or if a parent wanted to sleep close to an ill child), siblings tended to move in as well. Sleep can be challenging for parents of well children, and our studies show it’s even more so for parents of children who have experienced ALL.”
The researchers hope these studies increase awareness of maternal concerns after a child’s leukemia diagnosis and that leads to interventions to help mothers manage these lifestyle issues.

Credit: Bill Branson
The stress and lifestyle changes mothers experience after a child’s leukemia diagnosis do not disappear once the disease is in the maintenance phase, new research suggests.
One study showed that mothers of patients with acute lymphoblastic leukemia (ALL) continued to exhibit signs of stress, anxiety, and depression when their child’s disease was in the maintenance phase.
And another study showed that a family’s daily schedule and sleeping arrangements did not return to the way they were before the child’s diagnosis.
“Even though these mothers were in the maintenance phase of their child’s illness, and the prognosis was good, we heard them say over and over that things could never go back to what they were before,” said study author Madalynn Neu, PhD, RN, of the Colorado University College of Nursing.
Stress, depression, and anxiety
In the Journal of Pediatric Oncology Nursing, Dr Neu and her colleagues reported their analysis of stress, anxiety, and depression among the mothers of patients with ALL. The team evaluated 26 mothers of ALL patients and 26 mothers of healthy children, who were matched according to the child’s age and gender.
To assess anxiety, the researchers collected salivary cortisol from the mothers 4 times a day for 3 consecutive days. The mothers also completed questionnaires—the Hospital Anxiety and Depression Scale and the Perceived Stress Scale.
More mothers in the ALL group had questionnaire scores indicating clinical anxiety (46%) and depressive symptoms (27%). And there was a trend toward increased stress in the mothers of ALL patients.
However, the researchers were surprised to find that the mothers’ anxiety levels—as measured by salivary cortisol—were similar to the mothers of well children.
“This may have been affected by the fact that even the control group wasn’t without anxiety,” said study author Ellen Matthews, PhD, RN, of the Colorado University College of Nursing.
“Financial, marital, social, and career concerns mean that parents of young children experience anxiety even without ALL.”
The sleep-wake experience
In the Journal of Pediatric Nursing, the researchers described their assessment of the sleep-wake experience for mothers of ALL patients. The team conducted interviews with 20 mothers, using open-ended, semi-structured questions, and the answers were transcribed verbatim.
Two main themes emerged during these interviews. The first was dubbed, “It’s a whole new cancer world” and contained 4 subthemes: losing normality, being off-balance/insecure, juggling duties, and making transitions.
“Many [of the mothers] had lost their normal lives—lost jobs, houses, friends,” Dr Matthews said. “Some were juggling their time around their child’s needs, and they had fears about many things—fear of recurrence, fear of making a mistake with medication, fear their kids might get sick with an infection.”
The second theme that emerged in interviews was, “I don’t remember what it’s like to have sleep.” This also contained 4 subthemes: sleeping trouble before and after ALL, the child feeling sick at night, worrying, and coping with exhaustion.
The mothers also noted that once sleep arrangements had changed, they often did not return to their pretreatment “normal.”
“Mothers talked about the difficulty of sleep while giving steroid medication,” Dr Neu said. “And if the ill child got to stay up late watching movies, the siblings wanted to stay up too.”
“The same was true of sleeping in a parent’s room. If an ill child wanted to sleep close to a parent (or if a parent wanted to sleep close to an ill child), siblings tended to move in as well. Sleep can be challenging for parents of well children, and our studies show it’s even more so for parents of children who have experienced ALL.”
The researchers hope these studies increase awareness of maternal concerns after a child’s leukemia diagnosis and that leads to interventions to help mothers manage these lifestyle issues.

Credit: Bill Branson
The stress and lifestyle changes mothers experience after a child’s leukemia diagnosis do not disappear once the disease is in the maintenance phase, new research suggests.
One study showed that mothers of patients with acute lymphoblastic leukemia (ALL) continued to exhibit signs of stress, anxiety, and depression when their child’s disease was in the maintenance phase.
And another study showed that a family’s daily schedule and sleeping arrangements did not return to the way they were before the child’s diagnosis.
“Even though these mothers were in the maintenance phase of their child’s illness, and the prognosis was good, we heard them say over and over that things could never go back to what they were before,” said study author Madalynn Neu, PhD, RN, of the Colorado University College of Nursing.
Stress, depression, and anxiety
In the Journal of Pediatric Oncology Nursing, Dr Neu and her colleagues reported their analysis of stress, anxiety, and depression among the mothers of patients with ALL. The team evaluated 26 mothers of ALL patients and 26 mothers of healthy children, who were matched according to the child’s age and gender.
To assess anxiety, the researchers collected salivary cortisol from the mothers 4 times a day for 3 consecutive days. The mothers also completed questionnaires—the Hospital Anxiety and Depression Scale and the Perceived Stress Scale.
More mothers in the ALL group had questionnaire scores indicating clinical anxiety (46%) and depressive symptoms (27%). And there was a trend toward increased stress in the mothers of ALL patients.
However, the researchers were surprised to find that the mothers’ anxiety levels—as measured by salivary cortisol—were similar to the mothers of well children.
“This may have been affected by the fact that even the control group wasn’t without anxiety,” said study author Ellen Matthews, PhD, RN, of the Colorado University College of Nursing.
“Financial, marital, social, and career concerns mean that parents of young children experience anxiety even without ALL.”
The sleep-wake experience
In the Journal of Pediatric Nursing, the researchers described their assessment of the sleep-wake experience for mothers of ALL patients. The team conducted interviews with 20 mothers, using open-ended, semi-structured questions, and the answers were transcribed verbatim.
Two main themes emerged during these interviews. The first was dubbed, “It’s a whole new cancer world” and contained 4 subthemes: losing normality, being off-balance/insecure, juggling duties, and making transitions.
“Many [of the mothers] had lost their normal lives—lost jobs, houses, friends,” Dr Matthews said. “Some were juggling their time around their child’s needs, and they had fears about many things—fear of recurrence, fear of making a mistake with medication, fear their kids might get sick with an infection.”
The second theme that emerged in interviews was, “I don’t remember what it’s like to have sleep.” This also contained 4 subthemes: sleeping trouble before and after ALL, the child feeling sick at night, worrying, and coping with exhaustion.
The mothers also noted that once sleep arrangements had changed, they often did not return to their pretreatment “normal.”
“Mothers talked about the difficulty of sleep while giving steroid medication,” Dr Neu said. “And if the ill child got to stay up late watching movies, the siblings wanted to stay up too.”
“The same was true of sleeping in a parent’s room. If an ill child wanted to sleep close to a parent (or if a parent wanted to sleep close to an ill child), siblings tended to move in as well. Sleep can be challenging for parents of well children, and our studies show it’s even more so for parents of children who have experienced ALL.”
The researchers hope these studies increase awareness of maternal concerns after a child’s leukemia diagnosis and that leads to interventions to help mothers manage these lifestyle issues.
Targeting pathways can override resistance in ALL
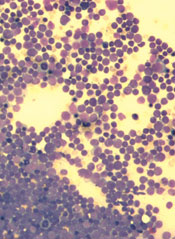
Inhibiting two biosynthesis pathways can override treatment resistance in acute lymphoblastic leukemia (ALL), preclinical research suggests.
Researchers found that inhibiting only one pathway did not effectively kill ALL cells. The cells simply used another pathway to replicate their DNA and continue dividing.
But inhibiting both pathways induced apoptosis in human leukemia cells and reduced tumor burden in mouse models of T- and B-cell ALL.
“This new, dual-targeting approach shows that we can overcome the redundancy in DNA synthesis in ALL cells and identifies a potential target for metabolic intervention in ALL, and possibly in other hematological cancers,” said study author Caius Radu, MD, of the University of California, Los Angeles.
He and his colleagues described this approach in The Journal of Experimental Medicine.
The research began with the knowledge that deoxyribonucleotide triphosphates, including deoxycytidine triphosphate (dCTP), are required for DNA replication, which is necessary for cell division. And dCTP can be produced by the de novo pathway or the nucleoside salvage pathway.
Dr Radu and his colleagues discovered that inhibiting the de novo pathway with the compound thymidine caused leukemia cells to switch to the nucleoside salvage pathway for dCTP production.
However, inhibiting both the de novo and nucleoside salvage pathways prevented dCTP production and proved lethal for leukemia cells.
A number of experiments elicited these results. In one, the researchers knocked down the deoxycytidine kinase (dCK) in human T-ALL cells to inhibit the nucleoside salvage pathway. Then, they administered thymidine to inhibit the de novo pathway. This resulted in dCTP depletion, stalled DNA replication, replication stress, DNA damage, and apoptosis.
The researchers also used the small-molecule inhibitor DI-39 to target dCK. They found that co-administration of DI-39 and thymidine induced replication stress and apoptosis in several leukemia cell lines: CEM, Jurkat, MOLT-4, NALM-6 and RS4;116.
The team then tested DI-39 and thymidine in mice bearing CEM tumors. They found the combination reduced tumor growth in mice bearing established, subcutaneous CEM xenografts and in mice with systemic T-ALL.
In the systemic T-ALL model, treatment with thymidine alone resulted in a 7-fold reduction in tumor burden compared to vehicle control or DI-39 alone. But when thymidine and DI-39 were administered together, mice saw a 100-fold reduction in tumor burden compared to thymidine alone.
The thymidine-DI-39 combination also proved effective against B-ALL cells and in a mouse model of B-ALL. However, the effects were not as great as those observed in T-ALL.
Finally, the researchers evaluated the effects of thymidine and DI-39 in hematopoietic progenitor cells. They looked at the Lineage- Sca-1+ c-Kit+ hematopoietic stem cell population, as well as short-term, long-term, and multipotent progenitor hematopoietic progenitor cells.
There was a minor decrease in the percentage of Lineage- Sca-1+ c-Kit+ cells after thymidine treatment. However, there were no other significant changes in progenitor cells between the treatment and control groups. Why leukemic cells and normal hematopoietic progenitors respond so differently to this treatment requires further investigation, the researchers said.
But they also noted that this study advances our understanding of DNA synthesis in leukemic cells and suggests that targeted metabolic intervention could be a new therapeutic approach in ALL.

Inhibiting two biosynthesis pathways can override treatment resistance in acute lymphoblastic leukemia (ALL), preclinical research suggests.
Researchers found that inhibiting only one pathway did not effectively kill ALL cells. The cells simply used another pathway to replicate their DNA and continue dividing.
But inhibiting both pathways induced apoptosis in human leukemia cells and reduced tumor burden in mouse models of T- and B-cell ALL.
“This new, dual-targeting approach shows that we can overcome the redundancy in DNA synthesis in ALL cells and identifies a potential target for metabolic intervention in ALL, and possibly in other hematological cancers,” said study author Caius Radu, MD, of the University of California, Los Angeles.
He and his colleagues described this approach in The Journal of Experimental Medicine.
The research began with the knowledge that deoxyribonucleotide triphosphates, including deoxycytidine triphosphate (dCTP), are required for DNA replication, which is necessary for cell division. And dCTP can be produced by the de novo pathway or the nucleoside salvage pathway.
Dr Radu and his colleagues discovered that inhibiting the de novo pathway with the compound thymidine caused leukemia cells to switch to the nucleoside salvage pathway for dCTP production.
However, inhibiting both the de novo and nucleoside salvage pathways prevented dCTP production and proved lethal for leukemia cells.
A number of experiments elicited these results. In one, the researchers knocked down the deoxycytidine kinase (dCK) in human T-ALL cells to inhibit the nucleoside salvage pathway. Then, they administered thymidine to inhibit the de novo pathway. This resulted in dCTP depletion, stalled DNA replication, replication stress, DNA damage, and apoptosis.
The researchers also used the small-molecule inhibitor DI-39 to target dCK. They found that co-administration of DI-39 and thymidine induced replication stress and apoptosis in several leukemia cell lines: CEM, Jurkat, MOLT-4, NALM-6 and RS4;116.
The team then tested DI-39 and thymidine in mice bearing CEM tumors. They found the combination reduced tumor growth in mice bearing established, subcutaneous CEM xenografts and in mice with systemic T-ALL.
In the systemic T-ALL model, treatment with thymidine alone resulted in a 7-fold reduction in tumor burden compared to vehicle control or DI-39 alone. But when thymidine and DI-39 were administered together, mice saw a 100-fold reduction in tumor burden compared to thymidine alone.
The thymidine-DI-39 combination also proved effective against B-ALL cells and in a mouse model of B-ALL. However, the effects were not as great as those observed in T-ALL.
Finally, the researchers evaluated the effects of thymidine and DI-39 in hematopoietic progenitor cells. They looked at the Lineage- Sca-1+ c-Kit+ hematopoietic stem cell population, as well as short-term, long-term, and multipotent progenitor hematopoietic progenitor cells.
There was a minor decrease in the percentage of Lineage- Sca-1+ c-Kit+ cells after thymidine treatment. However, there were no other significant changes in progenitor cells between the treatment and control groups. Why leukemic cells and normal hematopoietic progenitors respond so differently to this treatment requires further investigation, the researchers said.
But they also noted that this study advances our understanding of DNA synthesis in leukemic cells and suggests that targeted metabolic intervention could be a new therapeutic approach in ALL.

Inhibiting two biosynthesis pathways can override treatment resistance in acute lymphoblastic leukemia (ALL), preclinical research suggests.
Researchers found that inhibiting only one pathway did not effectively kill ALL cells. The cells simply used another pathway to replicate their DNA and continue dividing.
But inhibiting both pathways induced apoptosis in human leukemia cells and reduced tumor burden in mouse models of T- and B-cell ALL.
“This new, dual-targeting approach shows that we can overcome the redundancy in DNA synthesis in ALL cells and identifies a potential target for metabolic intervention in ALL, and possibly in other hematological cancers,” said study author Caius Radu, MD, of the University of California, Los Angeles.
He and his colleagues described this approach in The Journal of Experimental Medicine.
The research began with the knowledge that deoxyribonucleotide triphosphates, including deoxycytidine triphosphate (dCTP), are required for DNA replication, which is necessary for cell division. And dCTP can be produced by the de novo pathway or the nucleoside salvage pathway.
Dr Radu and his colleagues discovered that inhibiting the de novo pathway with the compound thymidine caused leukemia cells to switch to the nucleoside salvage pathway for dCTP production.
However, inhibiting both the de novo and nucleoside salvage pathways prevented dCTP production and proved lethal for leukemia cells.
A number of experiments elicited these results. In one, the researchers knocked down the deoxycytidine kinase (dCK) in human T-ALL cells to inhibit the nucleoside salvage pathway. Then, they administered thymidine to inhibit the de novo pathway. This resulted in dCTP depletion, stalled DNA replication, replication stress, DNA damage, and apoptosis.
The researchers also used the small-molecule inhibitor DI-39 to target dCK. They found that co-administration of DI-39 and thymidine induced replication stress and apoptosis in several leukemia cell lines: CEM, Jurkat, MOLT-4, NALM-6 and RS4;116.
The team then tested DI-39 and thymidine in mice bearing CEM tumors. They found the combination reduced tumor growth in mice bearing established, subcutaneous CEM xenografts and in mice with systemic T-ALL.
In the systemic T-ALL model, treatment with thymidine alone resulted in a 7-fold reduction in tumor burden compared to vehicle control or DI-39 alone. But when thymidine and DI-39 were administered together, mice saw a 100-fold reduction in tumor burden compared to thymidine alone.
The thymidine-DI-39 combination also proved effective against B-ALL cells and in a mouse model of B-ALL. However, the effects were not as great as those observed in T-ALL.
Finally, the researchers evaluated the effects of thymidine and DI-39 in hematopoietic progenitor cells. They looked at the Lineage- Sca-1+ c-Kit+ hematopoietic stem cell population, as well as short-term, long-term, and multipotent progenitor hematopoietic progenitor cells.
There was a minor decrease in the percentage of Lineage- Sca-1+ c-Kit+ cells after thymidine treatment. However, there were no other significant changes in progenitor cells between the treatment and control groups. Why leukemic cells and normal hematopoietic progenitors respond so differently to this treatment requires further investigation, the researchers said.
But they also noted that this study advances our understanding of DNA synthesis in leukemic cells and suggests that targeted metabolic intervention could be a new therapeutic approach in ALL.
Investigating the cause of infant leukemias
Infants who develop leukemia during the first year of life inherit a combination of genetic variations that can make them highly susceptible to the disease, according to a study published in Leukemia.
Results of whole-exome sequencing suggested that infants with leukemia inherited genetic variants from both parents that, by themselves, would not cause leukemia but, in combination, put the infants at high risk of developing the disease.
“We sequenced every single gene and found that infants with leukemia were born with an excess of damaging changes in genes known to be linked to leukemia,” said study author Todd Druley, MD, PhD, of Washington University School of Medicine in St Louis, Missouri.
“For each child, both parents carried a few harmful genetic variations in their DNA, and, just by chance, their child inherited all of these changes.”
However, it’s unlikely that the inherited variations alone cause leukemia, Dr Druley said. The infants likely needed to accumulate a few additional variations.
To uncover these findings, Dr Druley and his colleagues performed whole-exome sequencing in infants with acute myeloid leukemia (AML), infants with acute lymphoblastic leukemia (ALL), and the mothers of these children. The researchers used the process of elimination to determine a father’s contribution to a child’s DNA.
Among the 23 families studied, there was no history of pediatric cancers. As a comparison, the researchers also sequenced the DNA of 25 healthy children.
The team found the average amount of congenital coding variations was higher in infants with leukemia than in their mothers or the control subjects. The average total variants per exome was 1264.4 in infants with ALL, 1112.6 in their mothers, 2549.9 in infants with AML, 1225.0 in their mothers, and 582.8 in healthy controls.
The researchers then decided to home in on variants that were likely to impart a functional effect associated with leukemia. Using the COSMIC database, the team identified 126 ALL-associated genes and 655 AML-associated genes.
They found an average of 12.1 variants per ALL patient in the ALL-associated genes and 163.4 variants per AML patient in the AML-associated genes. There were 6.4 ALL-associated variants in the ALL patients’ mothers, 132.5 AML-associated variants in the AML patients’ mothers, 1.9 ALL variants in controls, and 27.5 AML variants in controls.
To prioritize genes that might be most relevant to infant leukemia, the researchers looked for compound heterozygous genes and the genes that were most commonly variant in all patients.
All of the infants with AML and 50% of the infants with ALL were compound heterozygotes for MLL3. Sixty-seven percent of AML patients were compound heterozygotes for RYR1 and FLG, and 50% of ALL patients were compound heterozygotes for RBMX.
The most variant (but not necessarily compound heterozygous) AML-associated genes in infants with AML were TTN, MLL3, and FLG. But the ALL-associated genes MDN1, SYNE1, and MLL2 were frequently variable in AML patients as well.
For infants with ALL, MDN1 was the most variable ALL-associated gene. But these infants also had frequent variations in the AML-associated genes TTN, RBMX, and MLL3.
Dr Druley and his colleagues plan to study these variations in more detail to understand how they contribute to infant leukemia development.
Infants who develop leukemia during the first year of life inherit a combination of genetic variations that can make them highly susceptible to the disease, according to a study published in Leukemia.
Results of whole-exome sequencing suggested that infants with leukemia inherited genetic variants from both parents that, by themselves, would not cause leukemia but, in combination, put the infants at high risk of developing the disease.
“We sequenced every single gene and found that infants with leukemia were born with an excess of damaging changes in genes known to be linked to leukemia,” said study author Todd Druley, MD, PhD, of Washington University School of Medicine in St Louis, Missouri.
“For each child, both parents carried a few harmful genetic variations in their DNA, and, just by chance, their child inherited all of these changes.”
However, it’s unlikely that the inherited variations alone cause leukemia, Dr Druley said. The infants likely needed to accumulate a few additional variations.
To uncover these findings, Dr Druley and his colleagues performed whole-exome sequencing in infants with acute myeloid leukemia (AML), infants with acute lymphoblastic leukemia (ALL), and the mothers of these children. The researchers used the process of elimination to determine a father’s contribution to a child’s DNA.
Among the 23 families studied, there was no history of pediatric cancers. As a comparison, the researchers also sequenced the DNA of 25 healthy children.
The team found the average amount of congenital coding variations was higher in infants with leukemia than in their mothers or the control subjects. The average total variants per exome was 1264.4 in infants with ALL, 1112.6 in their mothers, 2549.9 in infants with AML, 1225.0 in their mothers, and 582.8 in healthy controls.
The researchers then decided to home in on variants that were likely to impart a functional effect associated with leukemia. Using the COSMIC database, the team identified 126 ALL-associated genes and 655 AML-associated genes.
They found an average of 12.1 variants per ALL patient in the ALL-associated genes and 163.4 variants per AML patient in the AML-associated genes. There were 6.4 ALL-associated variants in the ALL patients’ mothers, 132.5 AML-associated variants in the AML patients’ mothers, 1.9 ALL variants in controls, and 27.5 AML variants in controls.
To prioritize genes that might be most relevant to infant leukemia, the researchers looked for compound heterozygous genes and the genes that were most commonly variant in all patients.
All of the infants with AML and 50% of the infants with ALL were compound heterozygotes for MLL3. Sixty-seven percent of AML patients were compound heterozygotes for RYR1 and FLG, and 50% of ALL patients were compound heterozygotes for RBMX.
The most variant (but not necessarily compound heterozygous) AML-associated genes in infants with AML were TTN, MLL3, and FLG. But the ALL-associated genes MDN1, SYNE1, and MLL2 were frequently variable in AML patients as well.
For infants with ALL, MDN1 was the most variable ALL-associated gene. But these infants also had frequent variations in the AML-associated genes TTN, RBMX, and MLL3.
Dr Druley and his colleagues plan to study these variations in more detail to understand how they contribute to infant leukemia development.
Infants who develop leukemia during the first year of life inherit a combination of genetic variations that can make them highly susceptible to the disease, according to a study published in Leukemia.
Results of whole-exome sequencing suggested that infants with leukemia inherited genetic variants from both parents that, by themselves, would not cause leukemia but, in combination, put the infants at high risk of developing the disease.
“We sequenced every single gene and found that infants with leukemia were born with an excess of damaging changes in genes known to be linked to leukemia,” said study author Todd Druley, MD, PhD, of Washington University School of Medicine in St Louis, Missouri.
“For each child, both parents carried a few harmful genetic variations in their DNA, and, just by chance, their child inherited all of these changes.”
However, it’s unlikely that the inherited variations alone cause leukemia, Dr Druley said. The infants likely needed to accumulate a few additional variations.
To uncover these findings, Dr Druley and his colleagues performed whole-exome sequencing in infants with acute myeloid leukemia (AML), infants with acute lymphoblastic leukemia (ALL), and the mothers of these children. The researchers used the process of elimination to determine a father’s contribution to a child’s DNA.
Among the 23 families studied, there was no history of pediatric cancers. As a comparison, the researchers also sequenced the DNA of 25 healthy children.
The team found the average amount of congenital coding variations was higher in infants with leukemia than in their mothers or the control subjects. The average total variants per exome was 1264.4 in infants with ALL, 1112.6 in their mothers, 2549.9 in infants with AML, 1225.0 in their mothers, and 582.8 in healthy controls.
The researchers then decided to home in on variants that were likely to impart a functional effect associated with leukemia. Using the COSMIC database, the team identified 126 ALL-associated genes and 655 AML-associated genes.
They found an average of 12.1 variants per ALL patient in the ALL-associated genes and 163.4 variants per AML patient in the AML-associated genes. There were 6.4 ALL-associated variants in the ALL patients’ mothers, 132.5 AML-associated variants in the AML patients’ mothers, 1.9 ALL variants in controls, and 27.5 AML variants in controls.
To prioritize genes that might be most relevant to infant leukemia, the researchers looked for compound heterozygous genes and the genes that were most commonly variant in all patients.
All of the infants with AML and 50% of the infants with ALL were compound heterozygotes for MLL3. Sixty-seven percent of AML patients were compound heterozygotes for RYR1 and FLG, and 50% of ALL patients were compound heterozygotes for RBMX.
The most variant (but not necessarily compound heterozygous) AML-associated genes in infants with AML were TTN, MLL3, and FLG. But the ALL-associated genes MDN1, SYNE1, and MLL2 were frequently variable in AML patients as well.
For infants with ALL, MDN1 was the most variable ALL-associated gene. But these infants also had frequent variations in the AML-associated genes TTN, RBMX, and MLL3.
Dr Druley and his colleagues plan to study these variations in more detail to understand how they contribute to infant leukemia development.