User login
ASCO updates guidelines on antiemetic use in cancer patients
The American Society of Clinical Oncology (ASCO) has updated its clinical practice guidelines on the use of antiemetics in cancer patients.
The update, published in the Journal of Clinical Oncology, provides new evidence-based information on the appropriate use of olanzapine, NK1 receptor antagonists, and dexamethasone.
“The adverse impact of inadequately controlled nausea and vomiting on patients’ quality of life is well documented,” said Paul J. Hesketh, MD, co-chair of the ASCO expert panel that updated the guidelines.
“By following the ASCO antiemetics guideline, clinicians have the opportunity to improve patients’ quality of life by minimizing treatment-induced emesis.”
To update ASCO’s guidelines on antiemetics, the expert panel conducted a systematic review of the medical literature published between November 2009 and June 2016. The panel included members with expertise in medical oncology, radiation oncology, nursing, pharmacy, and health services research, as well as a patient representative.
“Tremendous progress has been realized over the last 25 years in the prevention of chemotherapy-induced nausea and vomiting with the introduction of new classes of antiemetic agents,” said Mark G. Kris, MD, co-chair of the expert panel that updated the guidelines.
“The full benefit of these treatment advances will only be realized, however, if evidence-based guidelines are fully implemented.”
Key recommendations in the updated guidelines include:
For adults receiving chemotherapy with a high risk for nausea and vomiting (eg, cisplatin or the combination of cyclophosphamide and an anthracycline), olanzapine should be added to standard antiemetic regimens (the combination of a 5-HT3 receptor antagonist, an NK1 receptor antagonist, and dexamethasone). Olanzapine also helps individuals who experience symptoms despite receiving medicines to prevent vomiting before chemotherapy is given.
For adults receiving carboplatin-based chemotherapy or high-dose chemotherapy and children receiving chemotherapy with a high risk for nausea and vomiting, an NK1 receptor antagonist should be added to the standard antiemetic regimen (the combination of 5-HT3 receptor antagonist and dexamethasone).
Dexamethasone treatment can be limited to the day of chemotherapy administration in patients receiving an anthracycline and cyclophosphamide.
Dronabinol and nabilone, cannabinoids approved by the US Food and Drug Administration, can be used to treat nausea and vomiting that is resistant to standard antiemetic therapies. Evidence remains insufficient to recommend medical marijuana for either prevention or treatment of nausea and vomiting in patients with cancer receiving chemotherapy or radiation therapy. ![]()
The American Society of Clinical Oncology (ASCO) has updated its clinical practice guidelines on the use of antiemetics in cancer patients.
The update, published in the Journal of Clinical Oncology, provides new evidence-based information on the appropriate use of olanzapine, NK1 receptor antagonists, and dexamethasone.
“The adverse impact of inadequately controlled nausea and vomiting on patients’ quality of life is well documented,” said Paul J. Hesketh, MD, co-chair of the ASCO expert panel that updated the guidelines.
“By following the ASCO antiemetics guideline, clinicians have the opportunity to improve patients’ quality of life by minimizing treatment-induced emesis.”
To update ASCO’s guidelines on antiemetics, the expert panel conducted a systematic review of the medical literature published between November 2009 and June 2016. The panel included members with expertise in medical oncology, radiation oncology, nursing, pharmacy, and health services research, as well as a patient representative.
“Tremendous progress has been realized over the last 25 years in the prevention of chemotherapy-induced nausea and vomiting with the introduction of new classes of antiemetic agents,” said Mark G. Kris, MD, co-chair of the expert panel that updated the guidelines.
“The full benefit of these treatment advances will only be realized, however, if evidence-based guidelines are fully implemented.”
Key recommendations in the updated guidelines include:
For adults receiving chemotherapy with a high risk for nausea and vomiting (eg, cisplatin or the combination of cyclophosphamide and an anthracycline), olanzapine should be added to standard antiemetic regimens (the combination of a 5-HT3 receptor antagonist, an NK1 receptor antagonist, and dexamethasone). Olanzapine also helps individuals who experience symptoms despite receiving medicines to prevent vomiting before chemotherapy is given.
For adults receiving carboplatin-based chemotherapy or high-dose chemotherapy and children receiving chemotherapy with a high risk for nausea and vomiting, an NK1 receptor antagonist should be added to the standard antiemetic regimen (the combination of 5-HT3 receptor antagonist and dexamethasone).
Dexamethasone treatment can be limited to the day of chemotherapy administration in patients receiving an anthracycline and cyclophosphamide.
Dronabinol and nabilone, cannabinoids approved by the US Food and Drug Administration, can be used to treat nausea and vomiting that is resistant to standard antiemetic therapies. Evidence remains insufficient to recommend medical marijuana for either prevention or treatment of nausea and vomiting in patients with cancer receiving chemotherapy or radiation therapy. ![]()
The American Society of Clinical Oncology (ASCO) has updated its clinical practice guidelines on the use of antiemetics in cancer patients.
The update, published in the Journal of Clinical Oncology, provides new evidence-based information on the appropriate use of olanzapine, NK1 receptor antagonists, and dexamethasone.
“The adverse impact of inadequately controlled nausea and vomiting on patients’ quality of life is well documented,” said Paul J. Hesketh, MD, co-chair of the ASCO expert panel that updated the guidelines.
“By following the ASCO antiemetics guideline, clinicians have the opportunity to improve patients’ quality of life by minimizing treatment-induced emesis.”
To update ASCO’s guidelines on antiemetics, the expert panel conducted a systematic review of the medical literature published between November 2009 and June 2016. The panel included members with expertise in medical oncology, radiation oncology, nursing, pharmacy, and health services research, as well as a patient representative.
“Tremendous progress has been realized over the last 25 years in the prevention of chemotherapy-induced nausea and vomiting with the introduction of new classes of antiemetic agents,” said Mark G. Kris, MD, co-chair of the expert panel that updated the guidelines.
“The full benefit of these treatment advances will only be realized, however, if evidence-based guidelines are fully implemented.”
Key recommendations in the updated guidelines include:
For adults receiving chemotherapy with a high risk for nausea and vomiting (eg, cisplatin or the combination of cyclophosphamide and an anthracycline), olanzapine should be added to standard antiemetic regimens (the combination of a 5-HT3 receptor antagonist, an NK1 receptor antagonist, and dexamethasone). Olanzapine also helps individuals who experience symptoms despite receiving medicines to prevent vomiting before chemotherapy is given.
For adults receiving carboplatin-based chemotherapy or high-dose chemotherapy and children receiving chemotherapy with a high risk for nausea and vomiting, an NK1 receptor antagonist should be added to the standard antiemetic regimen (the combination of 5-HT3 receptor antagonist and dexamethasone).
Dexamethasone treatment can be limited to the day of chemotherapy administration in patients receiving an anthracycline and cyclophosphamide.
Dronabinol and nabilone, cannabinoids approved by the US Food and Drug Administration, can be used to treat nausea and vomiting that is resistant to standard antiemetic therapies. Evidence remains insufficient to recommend medical marijuana for either prevention or treatment of nausea and vomiting in patients with cancer receiving chemotherapy or radiation therapy. ![]()
Models provide new understanding of sickle cell disease
Computer models have revealed new details of what happens inside a red blood cell affected by sickle cell disease (SCD), according to research published in Biophysical Journal.
In patients with SCD, mutated hemoglobin can polymerize, assembling into long fibers that push against the membranes of red blood cells and force them out of shape.
“The goal of our work is to model both how these sickle hemoglobin fibers form as well as the mechanical properties of those fibers,” said study author Lu Lu, a PhD student at Brown University in Providence, Rhode Island.
“There had been separate models for each of these things individually developed by us, but this brings those together into one comprehensive model.”
The model uses detailed biomechanical data on how sickle hemoglobin molecules behave and bind with each other to simulate the assembly of a polymer fiber.
Prior to this work, the problem had been that, as the fiber grows, so does the amount of data the model must crunch. Modeling an entire polymer fiber at a cellular scale using the details of each molecule was simply too computationally expensive.
“Even the world’s fastest supercomputers wouldn’t be able to handle it,” said study author George Karniadakis, PhD, of Brown University.
“There’s just too much happening and no way to capture it all computationally. That’s what we were able to overcome with this work.”
The researchers’ solution was to apply what they call a mesoscopic adaptive resolution scheme (MARS).
The MARS model calculates the detailed dynamics of each individual hemoglobin molecule only at the end of polymer fibers, where new molecules are being recruited into the fiber.
Once 4 layers of a fiber have been established, the model automatically dials back the resolution at which it represents that section. The model retains the important information about how the fiber behaves mechanically but glosses over the fine details of each constituent molecule.
“By eliminating the fine details where we don’t need them, we develop a model that can simulate this whole process and its effects on a red blood cell,” Dr Karniadakis said.
Using the new MARS simulations, the researchers were able to show how different configurations of growing polymer fibers are able to produce cells with different shapes.
“We are able to produce a polymerization profile for each of the cell types associated with the disease,” Dr Karniadakis said. “Now, the goal is to use these models to look for ways of preventing the disease onset.”
Using these new models, Dr Karniadakis and his colleagues can run simulations that include fetal hemoglobin. Those simulations could be used to confirm the theory that fetal hemoglobin disrupts polymerization, as well as determine how much fetal hemoglobin is necessary.
That could help in establishing better dosage guidelines for hydroxyurea or in developing new and more effective drugs for SCD, according to the researchers.
“The models give us a way to do preliminary testing on new approaches to stopping this disease,” Dr Karniadakis said. “Now that we can simulate the entire polymerization process, we think the models will be much more useful.” ![]()
Computer models have revealed new details of what happens inside a red blood cell affected by sickle cell disease (SCD), according to research published in Biophysical Journal.
In patients with SCD, mutated hemoglobin can polymerize, assembling into long fibers that push against the membranes of red blood cells and force them out of shape.
“The goal of our work is to model both how these sickle hemoglobin fibers form as well as the mechanical properties of those fibers,” said study author Lu Lu, a PhD student at Brown University in Providence, Rhode Island.
“There had been separate models for each of these things individually developed by us, but this brings those together into one comprehensive model.”
The model uses detailed biomechanical data on how sickle hemoglobin molecules behave and bind with each other to simulate the assembly of a polymer fiber.
Prior to this work, the problem had been that, as the fiber grows, so does the amount of data the model must crunch. Modeling an entire polymer fiber at a cellular scale using the details of each molecule was simply too computationally expensive.
“Even the world’s fastest supercomputers wouldn’t be able to handle it,” said study author George Karniadakis, PhD, of Brown University.
“There’s just too much happening and no way to capture it all computationally. That’s what we were able to overcome with this work.”
The researchers’ solution was to apply what they call a mesoscopic adaptive resolution scheme (MARS).
The MARS model calculates the detailed dynamics of each individual hemoglobin molecule only at the end of polymer fibers, where new molecules are being recruited into the fiber.
Once 4 layers of a fiber have been established, the model automatically dials back the resolution at which it represents that section. The model retains the important information about how the fiber behaves mechanically but glosses over the fine details of each constituent molecule.
“By eliminating the fine details where we don’t need them, we develop a model that can simulate this whole process and its effects on a red blood cell,” Dr Karniadakis said.
Using the new MARS simulations, the researchers were able to show how different configurations of growing polymer fibers are able to produce cells with different shapes.
“We are able to produce a polymerization profile for each of the cell types associated with the disease,” Dr Karniadakis said. “Now, the goal is to use these models to look for ways of preventing the disease onset.”
Using these new models, Dr Karniadakis and his colleagues can run simulations that include fetal hemoglobin. Those simulations could be used to confirm the theory that fetal hemoglobin disrupts polymerization, as well as determine how much fetal hemoglobin is necessary.
That could help in establishing better dosage guidelines for hydroxyurea or in developing new and more effective drugs for SCD, according to the researchers.
“The models give us a way to do preliminary testing on new approaches to stopping this disease,” Dr Karniadakis said. “Now that we can simulate the entire polymerization process, we think the models will be much more useful.” ![]()
Computer models have revealed new details of what happens inside a red blood cell affected by sickle cell disease (SCD), according to research published in Biophysical Journal.
In patients with SCD, mutated hemoglobin can polymerize, assembling into long fibers that push against the membranes of red blood cells and force them out of shape.
“The goal of our work is to model both how these sickle hemoglobin fibers form as well as the mechanical properties of those fibers,” said study author Lu Lu, a PhD student at Brown University in Providence, Rhode Island.
“There had been separate models for each of these things individually developed by us, but this brings those together into one comprehensive model.”
The model uses detailed biomechanical data on how sickle hemoglobin molecules behave and bind with each other to simulate the assembly of a polymer fiber.
Prior to this work, the problem had been that, as the fiber grows, so does the amount of data the model must crunch. Modeling an entire polymer fiber at a cellular scale using the details of each molecule was simply too computationally expensive.
“Even the world’s fastest supercomputers wouldn’t be able to handle it,” said study author George Karniadakis, PhD, of Brown University.
“There’s just too much happening and no way to capture it all computationally. That’s what we were able to overcome with this work.”
The researchers’ solution was to apply what they call a mesoscopic adaptive resolution scheme (MARS).
The MARS model calculates the detailed dynamics of each individual hemoglobin molecule only at the end of polymer fibers, where new molecules are being recruited into the fiber.
Once 4 layers of a fiber have been established, the model automatically dials back the resolution at which it represents that section. The model retains the important information about how the fiber behaves mechanically but glosses over the fine details of each constituent molecule.
“By eliminating the fine details where we don’t need them, we develop a model that can simulate this whole process and its effects on a red blood cell,” Dr Karniadakis said.
Using the new MARS simulations, the researchers were able to show how different configurations of growing polymer fibers are able to produce cells with different shapes.
“We are able to produce a polymerization profile for each of the cell types associated with the disease,” Dr Karniadakis said. “Now, the goal is to use these models to look for ways of preventing the disease onset.”
Using these new models, Dr Karniadakis and his colleagues can run simulations that include fetal hemoglobin. Those simulations could be used to confirm the theory that fetal hemoglobin disrupts polymerization, as well as determine how much fetal hemoglobin is necessary.
That could help in establishing better dosage guidelines for hydroxyurea or in developing new and more effective drugs for SCD, according to the researchers.
“The models give us a way to do preliminary testing on new approaches to stopping this disease,” Dr Karniadakis said. “Now that we can simulate the entire polymerization process, we think the models will be much more useful.” ![]()
FDA grants drug breakthrough designation for AML
The US Food and Drug Administration (FDA) has granted breakthrough therapy designation to venetoclax (Venclexta®).
The designation is for venetoclax in combination with low-dose cytarabine to treat elderly patients with previously untreated acute myeloid leukemia (AML) who are ineligible for intensive chemotherapy.
The FDA’s breakthrough designation is intended to expedite the development and review of new treatments for serious or life-threatening conditions.
Breakthrough designation entitles the company developing a therapy to more intensive FDA guidance on an efficient and accelerated development program, as well as eligibility for other actions to expedite FDA review, such as a rolling submission and priority review.
To earn breakthrough designation, a treatment must show encouraging early clinical results demonstrating substantial improvement over available therapies with regard to a clinically significant endpoint, or it must fulfill an unmet need.
About venetoclax
Venetoclax is a small molecule designed to selectively bind and inhibit the BCL-2 protein. The drug is being developed by AbbVie and Roche.
Last year, the FDA granted venetoclax accelerated approval to treat patients with chronic lymphocytic leukemia who have 17p deletion and have received at least one prior therapy. Continued approval of venetoclax for this indication may be contingent upon verification of clinical benefit in confirmatory trials.
The FDA granted venetoclax breakthrough therapy designation for the AML indication based on data from an ongoing phase 1/2 study. Preliminary data from the study were presented at the 22nd European Hematology Association (EHA) Annual Congress.
The presentation included data on 61 elderly patients (older than 65) with previously untreated AML who were ineligible for intensive chemotherapy.
They received venetoclax in combination with low-dose cytarabine (as well as prophylaxis for tumor lysis syndrome). The patients’ median time on treatment was 6 months (range, <1 to 19 months), and 72% of patients discontinued treatment.
The overall response rate was 65%. Twenty-five percent of patients achieved a complete response, 38% had a complete response with incomplete blood count recovery, and 2% had a partial response.
The 30-day death rate was 3%, the 60-day death rate was 15%, and the median overall survival was approximately 12 months.
The most common adverse events of any grade (occurring in at least 30% of patients) were nausea (74%), hypokalemia (46%), diarrhea (46%), fatigue (44%), decreased appetite (41%), constipation (34%), hypomagnesemia (34%), vomiting (31%), thrombocytopenia (44%), febrile neutropenia (38%), and neutropenia (33%).
Grade 3/4 adverse events (occurring in at least 10% of patients) included thrombocytopenia (44%), febrile neutropenia (36%), neutropenia (33%), anemia (28%), hypokalemia (16%), hypophosphatemia (13%), and hypertension (12%). ![]()
The US Food and Drug Administration (FDA) has granted breakthrough therapy designation to venetoclax (Venclexta®).
The designation is for venetoclax in combination with low-dose cytarabine to treat elderly patients with previously untreated acute myeloid leukemia (AML) who are ineligible for intensive chemotherapy.
The FDA’s breakthrough designation is intended to expedite the development and review of new treatments for serious or life-threatening conditions.
Breakthrough designation entitles the company developing a therapy to more intensive FDA guidance on an efficient and accelerated development program, as well as eligibility for other actions to expedite FDA review, such as a rolling submission and priority review.
To earn breakthrough designation, a treatment must show encouraging early clinical results demonstrating substantial improvement over available therapies with regard to a clinically significant endpoint, or it must fulfill an unmet need.
About venetoclax
Venetoclax is a small molecule designed to selectively bind and inhibit the BCL-2 protein. The drug is being developed by AbbVie and Roche.
Last year, the FDA granted venetoclax accelerated approval to treat patients with chronic lymphocytic leukemia who have 17p deletion and have received at least one prior therapy. Continued approval of venetoclax for this indication may be contingent upon verification of clinical benefit in confirmatory trials.
The FDA granted venetoclax breakthrough therapy designation for the AML indication based on data from an ongoing phase 1/2 study. Preliminary data from the study were presented at the 22nd European Hematology Association (EHA) Annual Congress.
The presentation included data on 61 elderly patients (older than 65) with previously untreated AML who were ineligible for intensive chemotherapy.
They received venetoclax in combination with low-dose cytarabine (as well as prophylaxis for tumor lysis syndrome). The patients’ median time on treatment was 6 months (range, <1 to 19 months), and 72% of patients discontinued treatment.
The overall response rate was 65%. Twenty-five percent of patients achieved a complete response, 38% had a complete response with incomplete blood count recovery, and 2% had a partial response.
The 30-day death rate was 3%, the 60-day death rate was 15%, and the median overall survival was approximately 12 months.
The most common adverse events of any grade (occurring in at least 30% of patients) were nausea (74%), hypokalemia (46%), diarrhea (46%), fatigue (44%), decreased appetite (41%), constipation (34%), hypomagnesemia (34%), vomiting (31%), thrombocytopenia (44%), febrile neutropenia (38%), and neutropenia (33%).
Grade 3/4 adverse events (occurring in at least 10% of patients) included thrombocytopenia (44%), febrile neutropenia (36%), neutropenia (33%), anemia (28%), hypokalemia (16%), hypophosphatemia (13%), and hypertension (12%). ![]()
The US Food and Drug Administration (FDA) has granted breakthrough therapy designation to venetoclax (Venclexta®).
The designation is for venetoclax in combination with low-dose cytarabine to treat elderly patients with previously untreated acute myeloid leukemia (AML) who are ineligible for intensive chemotherapy.
The FDA’s breakthrough designation is intended to expedite the development and review of new treatments for serious or life-threatening conditions.
Breakthrough designation entitles the company developing a therapy to more intensive FDA guidance on an efficient and accelerated development program, as well as eligibility for other actions to expedite FDA review, such as a rolling submission and priority review.
To earn breakthrough designation, a treatment must show encouraging early clinical results demonstrating substantial improvement over available therapies with regard to a clinically significant endpoint, or it must fulfill an unmet need.
About venetoclax
Venetoclax is a small molecule designed to selectively bind and inhibit the BCL-2 protein. The drug is being developed by AbbVie and Roche.
Last year, the FDA granted venetoclax accelerated approval to treat patients with chronic lymphocytic leukemia who have 17p deletion and have received at least one prior therapy. Continued approval of venetoclax for this indication may be contingent upon verification of clinical benefit in confirmatory trials.
The FDA granted venetoclax breakthrough therapy designation for the AML indication based on data from an ongoing phase 1/2 study. Preliminary data from the study were presented at the 22nd European Hematology Association (EHA) Annual Congress.
The presentation included data on 61 elderly patients (older than 65) with previously untreated AML who were ineligible for intensive chemotherapy.
They received venetoclax in combination with low-dose cytarabine (as well as prophylaxis for tumor lysis syndrome). The patients’ median time on treatment was 6 months (range, <1 to 19 months), and 72% of patients discontinued treatment.
The overall response rate was 65%. Twenty-five percent of patients achieved a complete response, 38% had a complete response with incomplete blood count recovery, and 2% had a partial response.
The 30-day death rate was 3%, the 60-day death rate was 15%, and the median overall survival was approximately 12 months.
The most common adverse events of any grade (occurring in at least 30% of patients) were nausea (74%), hypokalemia (46%), diarrhea (46%), fatigue (44%), decreased appetite (41%), constipation (34%), hypomagnesemia (34%), vomiting (31%), thrombocytopenia (44%), febrile neutropenia (38%), and neutropenia (33%).
Grade 3/4 adverse events (occurring in at least 10% of patients) included thrombocytopenia (44%), febrile neutropenia (36%), neutropenia (33%), anemia (28%), hypokalemia (16%), hypophosphatemia (13%), and hypertension (12%). ![]()
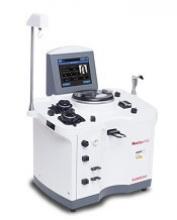
FDA clears new plasmapheresis system
The US Food and Drug Administration (FDA) has granted 510(k) clearance for Haemonetics Corporation’s NexSys PCS™ plasmapheresis system.
The open architecture of NexSys PCS facilitates bi-directional connectivity to donor management systems, enabling automated collection procedure programming and automated end-of-procedure documentation.
The guided operation, large touch screen, and on-screen troubleshooting assistance on NexSys PCS are designed to improve plasma center efficiency. The goal is to reduce donors’ time in centers and improve the centers’ collection capacity.
“NexSys PCS is designed to increase productivity and improve quality and compliance in plasma collection centers,” said Christopher Simon, CEO of Haemonetics.
“Each of these benefits is noteworthy and, when combined, we believe will unlock meaningful value for our customers.”
Haemonetics plans to begin limited production of NexSys PCS devices immediately and to pursue further regulatory clearances for additional enhancements to the overall product offering.
NexSys PCS was previously referred to as PCS 300. ![]()
The US Food and Drug Administration (FDA) has granted 510(k) clearance for Haemonetics Corporation’s NexSys PCS™ plasmapheresis system.
The open architecture of NexSys PCS facilitates bi-directional connectivity to donor management systems, enabling automated collection procedure programming and automated end-of-procedure documentation.
The guided operation, large touch screen, and on-screen troubleshooting assistance on NexSys PCS are designed to improve plasma center efficiency. The goal is to reduce donors’ time in centers and improve the centers’ collection capacity.
“NexSys PCS is designed to increase productivity and improve quality and compliance in plasma collection centers,” said Christopher Simon, CEO of Haemonetics.
“Each of these benefits is noteworthy and, when combined, we believe will unlock meaningful value for our customers.”
Haemonetics plans to begin limited production of NexSys PCS devices immediately and to pursue further regulatory clearances for additional enhancements to the overall product offering.
NexSys PCS was previously referred to as PCS 300. ![]()
The US Food and Drug Administration (FDA) has granted 510(k) clearance for Haemonetics Corporation’s NexSys PCS™ plasmapheresis system.
The open architecture of NexSys PCS facilitates bi-directional connectivity to donor management systems, enabling automated collection procedure programming and automated end-of-procedure documentation.
The guided operation, large touch screen, and on-screen troubleshooting assistance on NexSys PCS are designed to improve plasma center efficiency. The goal is to reduce donors’ time in centers and improve the centers’ collection capacity.
“NexSys PCS is designed to increase productivity and improve quality and compliance in plasma collection centers,” said Christopher Simon, CEO of Haemonetics.
“Each of these benefits is noteworthy and, when combined, we believe will unlock meaningful value for our customers.”
Haemonetics plans to begin limited production of NexSys PCS devices immediately and to pursue further regulatory clearances for additional enhancements to the overall product offering.
NexSys PCS was previously referred to as PCS 300. ![]()
CRISPR sheds light on dyskeratosis congenita
Gene editing has revealed how dyskeratosis congenita (DC) impairs the formation of blood cells, according to research published in Stem Cell Reports.
The discovery has opened up new lines of investigation into how to treat DC, which is characterized by shortened telomeres.
“Lengthening telomeres seems like a logical way to help these patients, but it could possibly come with its own set of problems,” said study author Luis F.Z. Batista, PhD, of the Washington University School of Medicine in St. Louis, Missouri.
“We would worry about encouraging cancer formation, for example, as high levels of the protein that lengthens telomeres—telomerase—are commonly found with cancer. But if we could find a way to block the signaling pathways that short telomeres activate—that specifically lead to the problems in blood cell formation—it could allow these patients to continue making blood cells.”
With this in mind, Dr Batista and his colleagues used CRISPR to edit into human embryonic stem cells a pair of mutations associated with DC— DKC1_A353V and TERT_P704S. These cells reproduced the telomere-shortening defect seen in patients with DC.
With this model, the researchers showed how the telomere defect leads to the gradual loss of blood cell formation and how blocking the downstream effects of the defect can reverse this loss, leading to normal production of blood cells.
Blocking this signaling pathway did not lengthen telomeres or stop their shortening, but it allowed the manufacturing of different types of blood cells to continue.
The researchers also made a discovery that provides a distinction regarding the detrimental effect of short telomeres during early development. The team found the defect did not hinder primitive hematopoiesis, but it did impair definitive hematopoiesis.
“This was tremendously interesting from a developmental biology perspective as well as from a disease modeling perspective,” said study author Christopher M. Sturgeon, PhD, of the Washington University School of Medicine. “We now have a platform to really dig deeper into understanding the mechanisms behind some forms of bone marrow failure.”
The researchers implicated high levels of the protein p53 as one of the signals that leads to the drop in definitive hematopoiesis.
“P53 is thought of as a guardian of the genome,” Dr Batista noted. “Mutations that disable p53 are associated with different types of cancer. Because of this, we would not consider directly trying to block p53 in these patients.”
“But what this study provides is proof-of-concept that this pathway is involved in this response. So we now are looking for ways to block the pathway further downstream without necessarily blocking p53 directly.”
Drs Batista and Sturgeon recently received a grant from the Department of Defense to investigate the pathway. The pair believes the strategy used in this study could be relevant for other bone marrow failure syndromes as well, such as Fanconi anemia and aplastic anemia. ![]()
Gene editing has revealed how dyskeratosis congenita (DC) impairs the formation of blood cells, according to research published in Stem Cell Reports.
The discovery has opened up new lines of investigation into how to treat DC, which is characterized by shortened telomeres.
“Lengthening telomeres seems like a logical way to help these patients, but it could possibly come with its own set of problems,” said study author Luis F.Z. Batista, PhD, of the Washington University School of Medicine in St. Louis, Missouri.
“We would worry about encouraging cancer formation, for example, as high levels of the protein that lengthens telomeres—telomerase—are commonly found with cancer. But if we could find a way to block the signaling pathways that short telomeres activate—that specifically lead to the problems in blood cell formation—it could allow these patients to continue making blood cells.”
With this in mind, Dr Batista and his colleagues used CRISPR to edit into human embryonic stem cells a pair of mutations associated with DC— DKC1_A353V and TERT_P704S. These cells reproduced the telomere-shortening defect seen in patients with DC.
With this model, the researchers showed how the telomere defect leads to the gradual loss of blood cell formation and how blocking the downstream effects of the defect can reverse this loss, leading to normal production of blood cells.
Blocking this signaling pathway did not lengthen telomeres or stop their shortening, but it allowed the manufacturing of different types of blood cells to continue.
The researchers also made a discovery that provides a distinction regarding the detrimental effect of short telomeres during early development. The team found the defect did not hinder primitive hematopoiesis, but it did impair definitive hematopoiesis.
“This was tremendously interesting from a developmental biology perspective as well as from a disease modeling perspective,” said study author Christopher M. Sturgeon, PhD, of the Washington University School of Medicine. “We now have a platform to really dig deeper into understanding the mechanisms behind some forms of bone marrow failure.”
The researchers implicated high levels of the protein p53 as one of the signals that leads to the drop in definitive hematopoiesis.
“P53 is thought of as a guardian of the genome,” Dr Batista noted. “Mutations that disable p53 are associated with different types of cancer. Because of this, we would not consider directly trying to block p53 in these patients.”
“But what this study provides is proof-of-concept that this pathway is involved in this response. So we now are looking for ways to block the pathway further downstream without necessarily blocking p53 directly.”
Drs Batista and Sturgeon recently received a grant from the Department of Defense to investigate the pathway. The pair believes the strategy used in this study could be relevant for other bone marrow failure syndromes as well, such as Fanconi anemia and aplastic anemia. ![]()
Gene editing has revealed how dyskeratosis congenita (DC) impairs the formation of blood cells, according to research published in Stem Cell Reports.
The discovery has opened up new lines of investigation into how to treat DC, which is characterized by shortened telomeres.
“Lengthening telomeres seems like a logical way to help these patients, but it could possibly come with its own set of problems,” said study author Luis F.Z. Batista, PhD, of the Washington University School of Medicine in St. Louis, Missouri.
“We would worry about encouraging cancer formation, for example, as high levels of the protein that lengthens telomeres—telomerase—are commonly found with cancer. But if we could find a way to block the signaling pathways that short telomeres activate—that specifically lead to the problems in blood cell formation—it could allow these patients to continue making blood cells.”
With this in mind, Dr Batista and his colleagues used CRISPR to edit into human embryonic stem cells a pair of mutations associated with DC— DKC1_A353V and TERT_P704S. These cells reproduced the telomere-shortening defect seen in patients with DC.
With this model, the researchers showed how the telomere defect leads to the gradual loss of blood cell formation and how blocking the downstream effects of the defect can reverse this loss, leading to normal production of blood cells.
Blocking this signaling pathway did not lengthen telomeres or stop their shortening, but it allowed the manufacturing of different types of blood cells to continue.
The researchers also made a discovery that provides a distinction regarding the detrimental effect of short telomeres during early development. The team found the defect did not hinder primitive hematopoiesis, but it did impair definitive hematopoiesis.
“This was tremendously interesting from a developmental biology perspective as well as from a disease modeling perspective,” said study author Christopher M. Sturgeon, PhD, of the Washington University School of Medicine. “We now have a platform to really dig deeper into understanding the mechanisms behind some forms of bone marrow failure.”
The researchers implicated high levels of the protein p53 as one of the signals that leads to the drop in definitive hematopoiesis.
“P53 is thought of as a guardian of the genome,” Dr Batista noted. “Mutations that disable p53 are associated with different types of cancer. Because of this, we would not consider directly trying to block p53 in these patients.”
“But what this study provides is proof-of-concept that this pathway is involved in this response. So we now are looking for ways to block the pathway further downstream without necessarily blocking p53 directly.”
Drs Batista and Sturgeon recently received a grant from the Department of Defense to investigate the pathway. The pair believes the strategy used in this study could be relevant for other bone marrow failure syndromes as well, such as Fanconi anemia and aplastic anemia. ![]()
Developing better mouse models
Researchers say they have developed a new approach to model human immune variation that overcomes the limitations of traditional mouse models.
With this approach, the team identified genetic markers that directly correlate with the outcome of inflammatory and malignant diseases in humans, including chronic lymphocytic leukemia and Burkitt lymphoma.
The findings suggest that accounting for immune diversity is critical to the success of predicting disease outcomes based on immune cell measurements.
The team reported these findings in Nature Communications.
Traditionally, researchers have relied on inbred mouse strains to gain insight into human diseases while reducing experimental noise.
“If you take a black, a brown, or a white mouse, each one will give you a different answer in the same assay,” said Klaus Ley, MD, of La Jolla Institute for Allergy and Immunology in California.
“For example, if you vaccinate them, their responses will be different, which creates a lot of experimental noise. However, when you think about patients, or even healthy people, we are all different.”
To mine those differences for information, the researchers embraced the experimental noise. Instead of analyzing a single inbred mouse strain, they turned to the hybrid mouse diversity panel (HDMP).
The HDMP is a panel of about 100 different inbred mouse strains that mirror the breadth of genetic and immunological diversity found in the human population.
The researchers studied the natural variation in the activation pattern of abdominal macrophages. Macrophages isolated from 83 different mouse strains from the HDMP were exposed to lipopolysaccharide (LPS), a major component of the outer wall of gram-negative bacteria.
“Fundamentally, when the immune system is confronted with gram-negative bacteria, it can deal with the situation in 2 ways—either it gets very angry and tries to kill the bacteria or it can wall them off in an attempt to live with it,” explained Dr Ley. “Both strategies carry a certain risk, but a long evolutionary history has ensured that mice and people can survive with either strategy.”
The LPS-induced reactions of the macrophages covered the whole spectrum—from very aggressive (LPS+) to very tolerant (LPS-), depending on the mouse strain.
Next, the researchers asked which genes were active during each type of response to identify gene signatures that correlated with LPS responsiveness.
The team then ran these gene signatures across various human gene expression data sets and discovered they strongly correlated with human disease outcomes.
For example, macrophages isolated from healthy joints were enriched in LPS-tolerant genes, whereas macrophages from rheumatoid arthritis patients were strongly skewed toward LPS-aggressive.
Since it was known that mice and people with the aggressive phenotype are better at fighting cancer, the researchers specifically asked whether the level of LPS responsiveness could predict tumor survival.
After analyzing data from 18,000 biopsies across 39 different tumor types, the team found the LPS+ gene signature strongly correlated with survival, while the LPS- signature correlated with cancer death.
The pattern was significant across many types of cancer, including chronic lymphocytic leukemia, Burkitt lymphoma, osteosarcoma, melanoma, and large-cell lung carcinoma. ![]()
Researchers say they have developed a new approach to model human immune variation that overcomes the limitations of traditional mouse models.
With this approach, the team identified genetic markers that directly correlate with the outcome of inflammatory and malignant diseases in humans, including chronic lymphocytic leukemia and Burkitt lymphoma.
The findings suggest that accounting for immune diversity is critical to the success of predicting disease outcomes based on immune cell measurements.
The team reported these findings in Nature Communications.
Traditionally, researchers have relied on inbred mouse strains to gain insight into human diseases while reducing experimental noise.
“If you take a black, a brown, or a white mouse, each one will give you a different answer in the same assay,” said Klaus Ley, MD, of La Jolla Institute for Allergy and Immunology in California.
“For example, if you vaccinate them, their responses will be different, which creates a lot of experimental noise. However, when you think about patients, or even healthy people, we are all different.”
To mine those differences for information, the researchers embraced the experimental noise. Instead of analyzing a single inbred mouse strain, they turned to the hybrid mouse diversity panel (HDMP).
The HDMP is a panel of about 100 different inbred mouse strains that mirror the breadth of genetic and immunological diversity found in the human population.
The researchers studied the natural variation in the activation pattern of abdominal macrophages. Macrophages isolated from 83 different mouse strains from the HDMP were exposed to lipopolysaccharide (LPS), a major component of the outer wall of gram-negative bacteria.
“Fundamentally, when the immune system is confronted with gram-negative bacteria, it can deal with the situation in 2 ways—either it gets very angry and tries to kill the bacteria or it can wall them off in an attempt to live with it,” explained Dr Ley. “Both strategies carry a certain risk, but a long evolutionary history has ensured that mice and people can survive with either strategy.”
The LPS-induced reactions of the macrophages covered the whole spectrum—from very aggressive (LPS+) to very tolerant (LPS-), depending on the mouse strain.
Next, the researchers asked which genes were active during each type of response to identify gene signatures that correlated with LPS responsiveness.
The team then ran these gene signatures across various human gene expression data sets and discovered they strongly correlated with human disease outcomes.
For example, macrophages isolated from healthy joints were enriched in LPS-tolerant genes, whereas macrophages from rheumatoid arthritis patients were strongly skewed toward LPS-aggressive.
Since it was known that mice and people with the aggressive phenotype are better at fighting cancer, the researchers specifically asked whether the level of LPS responsiveness could predict tumor survival.
After analyzing data from 18,000 biopsies across 39 different tumor types, the team found the LPS+ gene signature strongly correlated with survival, while the LPS- signature correlated with cancer death.
The pattern was significant across many types of cancer, including chronic lymphocytic leukemia, Burkitt lymphoma, osteosarcoma, melanoma, and large-cell lung carcinoma. ![]()
Researchers say they have developed a new approach to model human immune variation that overcomes the limitations of traditional mouse models.
With this approach, the team identified genetic markers that directly correlate with the outcome of inflammatory and malignant diseases in humans, including chronic lymphocytic leukemia and Burkitt lymphoma.
The findings suggest that accounting for immune diversity is critical to the success of predicting disease outcomes based on immune cell measurements.
The team reported these findings in Nature Communications.
Traditionally, researchers have relied on inbred mouse strains to gain insight into human diseases while reducing experimental noise.
“If you take a black, a brown, or a white mouse, each one will give you a different answer in the same assay,” said Klaus Ley, MD, of La Jolla Institute for Allergy and Immunology in California.
“For example, if you vaccinate them, their responses will be different, which creates a lot of experimental noise. However, when you think about patients, or even healthy people, we are all different.”
To mine those differences for information, the researchers embraced the experimental noise. Instead of analyzing a single inbred mouse strain, they turned to the hybrid mouse diversity panel (HDMP).
The HDMP is a panel of about 100 different inbred mouse strains that mirror the breadth of genetic and immunological diversity found in the human population.
The researchers studied the natural variation in the activation pattern of abdominal macrophages. Macrophages isolated from 83 different mouse strains from the HDMP were exposed to lipopolysaccharide (LPS), a major component of the outer wall of gram-negative bacteria.
“Fundamentally, when the immune system is confronted with gram-negative bacteria, it can deal with the situation in 2 ways—either it gets very angry and tries to kill the bacteria or it can wall them off in an attempt to live with it,” explained Dr Ley. “Both strategies carry a certain risk, but a long evolutionary history has ensured that mice and people can survive with either strategy.”
The LPS-induced reactions of the macrophages covered the whole spectrum—from very aggressive (LPS+) to very tolerant (LPS-), depending on the mouse strain.
Next, the researchers asked which genes were active during each type of response to identify gene signatures that correlated with LPS responsiveness.
The team then ran these gene signatures across various human gene expression data sets and discovered they strongly correlated with human disease outcomes.
For example, macrophages isolated from healthy joints were enriched in LPS-tolerant genes, whereas macrophages from rheumatoid arthritis patients were strongly skewed toward LPS-aggressive.
Since it was known that mice and people with the aggressive phenotype are better at fighting cancer, the researchers specifically asked whether the level of LPS responsiveness could predict tumor survival.
After analyzing data from 18,000 biopsies across 39 different tumor types, the team found the LPS+ gene signature strongly correlated with survival, while the LPS- signature correlated with cancer death.
The pattern was significant across many types of cancer, including chronic lymphocytic leukemia, Burkitt lymphoma, osteosarcoma, melanoma, and large-cell lung carcinoma. ![]()
Team makes ‘fundamental’ AML discovery
A newly identified pathway plays a “fundamental” role in acute myeloid leukemia (AML), according to researchers.
The team discovered that AML cells have a secretory pathway that leads to the production and release of the immune receptor Tim-3 and its ligand galectin-9, both of which prevent natural killer (NK) and other cytotoxic cells from killing the AML cells.
Vadim Sumbayev, PhD, of the University of Kent in the UK, and his colleagues recounted these findings in EBioMedicine.
The researchers found that AML cells—but not healthy blood cells—express a receptor called latrophilin 1 (LPHN1). LPHN1 induces activation of PKCα, which triggers the translation and secretion of Tim-3 and galectin-9.
Soluble Tim-3 prevents the secretion of interleukin 2, which is required for the activation of NK cells and cytotoxic T cells. Galectin-9 impairs the AML-cell-killing ability of NK cells and other cytotoxic lymphocytes.
The researchers said their work revealed both biomarkers for AML diagnostics and potential targets for AML treatment.
“Targeting this pathway will crucially enhance patients’ own immune defenses, helping them to eliminate leukemia cells,” Dr Sumbayev said.
He added that his group’s discovery might be applied to the treatment of other cancers as well. ![]()
A newly identified pathway plays a “fundamental” role in acute myeloid leukemia (AML), according to researchers.
The team discovered that AML cells have a secretory pathway that leads to the production and release of the immune receptor Tim-3 and its ligand galectin-9, both of which prevent natural killer (NK) and other cytotoxic cells from killing the AML cells.
Vadim Sumbayev, PhD, of the University of Kent in the UK, and his colleagues recounted these findings in EBioMedicine.
The researchers found that AML cells—but not healthy blood cells—express a receptor called latrophilin 1 (LPHN1). LPHN1 induces activation of PKCα, which triggers the translation and secretion of Tim-3 and galectin-9.
Soluble Tim-3 prevents the secretion of interleukin 2, which is required for the activation of NK cells and cytotoxic T cells. Galectin-9 impairs the AML-cell-killing ability of NK cells and other cytotoxic lymphocytes.
The researchers said their work revealed both biomarkers for AML diagnostics and potential targets for AML treatment.
“Targeting this pathway will crucially enhance patients’ own immune defenses, helping them to eliminate leukemia cells,” Dr Sumbayev said.
He added that his group’s discovery might be applied to the treatment of other cancers as well. ![]()
A newly identified pathway plays a “fundamental” role in acute myeloid leukemia (AML), according to researchers.
The team discovered that AML cells have a secretory pathway that leads to the production and release of the immune receptor Tim-3 and its ligand galectin-9, both of which prevent natural killer (NK) and other cytotoxic cells from killing the AML cells.
Vadim Sumbayev, PhD, of the University of Kent in the UK, and his colleagues recounted these findings in EBioMedicine.
The researchers found that AML cells—but not healthy blood cells—express a receptor called latrophilin 1 (LPHN1). LPHN1 induces activation of PKCα, which triggers the translation and secretion of Tim-3 and galectin-9.
Soluble Tim-3 prevents the secretion of interleukin 2, which is required for the activation of NK cells and cytotoxic T cells. Galectin-9 impairs the AML-cell-killing ability of NK cells and other cytotoxic lymphocytes.
The researchers said their work revealed both biomarkers for AML diagnostics and potential targets for AML treatment.
“Targeting this pathway will crucially enhance patients’ own immune defenses, helping them to eliminate leukemia cells,” Dr Sumbayev said.
He added that his group’s discovery might be applied to the treatment of other cancers as well.
Decreasing RBC transfusions saves center $1 million
Researchers have reported that interventions designed to decrease the need for red blood cell (RBC) transfusions proved effective when implemented at an academic medical center, and they resulted in an annual cost savings of more than $1 million.
The interventions included educational tools, a “best practice advisory,” and changes to the computerized provider order entry system.
They enabled the medical center to reduce multi-unit transfusions by 67.1% and transfusions for patients with hemoglobin (Hb) values of at least 7 g/dL by 47.4%.
Ian Jenkins, MD, of University of California San Diego (UCSD) Health, and his colleagues reported these results in The Joint Commission Journal on Quality and Patient Safety.
This work began with a multidisciplinary team at UCSD Health reviewing the transfusion literature on clinical trials, meta-analyses, guidelines, and improvement efforts.
The team used the information gleaned from this review and implemented the following interventions in an attempt to reduce unnecessary RBC transfusions:
- Educational tools (handouts, a video, PowerPoint presentations, etc)
- Enhancements to the health system’s computerized provider order entry system (eg, changing default RBC dose from 2 units to 1 unit)
- A “best practice advisory” intended to reduce unnecessary blood product use and costs by using real-time clinical decision support, a process for providing information at the point of care to help inform decisions about a patient’s care.
To assess the impact of their interventions, the researchers evaluated data on most non-infant, inpatient RBC transfusions given at UCSD Health between January 1, 2014, and September 30, 2016.
They excluded units given to patients with gastrointestinal bleeding and units given within 12 hours of a surgical procedure. The team said they excluded these transfusions because of the higher probability of unstable blood volume or special circumstances that might justify off-protocol transfusion.
The researchers then calculated the rate of inpatient RBC units transfused per 1000 patient-days (without exclusions), the percentage of inpatient RBC units transfused for patients with Hb ≥ 7 g/dL, and the percentage of multi-unit RBC transfusions, divided into 3 periods:
- Pre-intervention or baseline (January 1, 2014–September 30, 2014)
- During the intervention (October 1, 2014–April 30, 2015)
- Post-intervention (May 1, 2015–September 30, 2016).
There were 36,386 RBC units administered during the study period, which was 464,424 patient days.
Multi-unit transfusions decreased from 59.9% at baseline to 41.7% during the intervention period and 19.7% during the post-intervention period (P<0.0001).
Transfusions in patients with Hb values ≥ 7 g/dL decreased from 72.3% at baseline to 57.8% during the intervention period and 38.0% during the post-intervention period (P<0.0001).
The total RBC transfusion rate (units per 1000 patient-days) decreased from 89.8 at baseline to 78.1 during the intervention period and 72.7 during the post-intervention period (P<0.0001).
The estimated savings, based on a cost of $250 per RBC unit, was about $1,050,750 per year.
Researchers have reported that interventions designed to decrease the need for red blood cell (RBC) transfusions proved effective when implemented at an academic medical center, and they resulted in an annual cost savings of more than $1 million.
The interventions included educational tools, a “best practice advisory,” and changes to the computerized provider order entry system.
They enabled the medical center to reduce multi-unit transfusions by 67.1% and transfusions for patients with hemoglobin (Hb) values of at least 7 g/dL by 47.4%.
Ian Jenkins, MD, of University of California San Diego (UCSD) Health, and his colleagues reported these results in The Joint Commission Journal on Quality and Patient Safety.
This work began with a multidisciplinary team at UCSD Health reviewing the transfusion literature on clinical trials, meta-analyses, guidelines, and improvement efforts.
The team used the information gleaned from this review and implemented the following interventions in an attempt to reduce unnecessary RBC transfusions:
- Educational tools (handouts, a video, PowerPoint presentations, etc)
- Enhancements to the health system’s computerized provider order entry system (eg, changing default RBC dose from 2 units to 1 unit)
- A “best practice advisory” intended to reduce unnecessary blood product use and costs by using real-time clinical decision support, a process for providing information at the point of care to help inform decisions about a patient’s care.
To assess the impact of their interventions, the researchers evaluated data on most non-infant, inpatient RBC transfusions given at UCSD Health between January 1, 2014, and September 30, 2016.
They excluded units given to patients with gastrointestinal bleeding and units given within 12 hours of a surgical procedure. The team said they excluded these transfusions because of the higher probability of unstable blood volume or special circumstances that might justify off-protocol transfusion.
The researchers then calculated the rate of inpatient RBC units transfused per 1000 patient-days (without exclusions), the percentage of inpatient RBC units transfused for patients with Hb ≥ 7 g/dL, and the percentage of multi-unit RBC transfusions, divided into 3 periods:
- Pre-intervention or baseline (January 1, 2014–September 30, 2014)
- During the intervention (October 1, 2014–April 30, 2015)
- Post-intervention (May 1, 2015–September 30, 2016).
There were 36,386 RBC units administered during the study period, which was 464,424 patient days.
Multi-unit transfusions decreased from 59.9% at baseline to 41.7% during the intervention period and 19.7% during the post-intervention period (P<0.0001).
Transfusions in patients with Hb values ≥ 7 g/dL decreased from 72.3% at baseline to 57.8% during the intervention period and 38.0% during the post-intervention period (P<0.0001).
The total RBC transfusion rate (units per 1000 patient-days) decreased from 89.8 at baseline to 78.1 during the intervention period and 72.7 during the post-intervention period (P<0.0001).
The estimated savings, based on a cost of $250 per RBC unit, was about $1,050,750 per year.
Researchers have reported that interventions designed to decrease the need for red blood cell (RBC) transfusions proved effective when implemented at an academic medical center, and they resulted in an annual cost savings of more than $1 million.
The interventions included educational tools, a “best practice advisory,” and changes to the computerized provider order entry system.
They enabled the medical center to reduce multi-unit transfusions by 67.1% and transfusions for patients with hemoglobin (Hb) values of at least 7 g/dL by 47.4%.
Ian Jenkins, MD, of University of California San Diego (UCSD) Health, and his colleagues reported these results in The Joint Commission Journal on Quality and Patient Safety.
This work began with a multidisciplinary team at UCSD Health reviewing the transfusion literature on clinical trials, meta-analyses, guidelines, and improvement efforts.
The team used the information gleaned from this review and implemented the following interventions in an attempt to reduce unnecessary RBC transfusions:
- Educational tools (handouts, a video, PowerPoint presentations, etc)
- Enhancements to the health system’s computerized provider order entry system (eg, changing default RBC dose from 2 units to 1 unit)
- A “best practice advisory” intended to reduce unnecessary blood product use and costs by using real-time clinical decision support, a process for providing information at the point of care to help inform decisions about a patient’s care.
To assess the impact of their interventions, the researchers evaluated data on most non-infant, inpatient RBC transfusions given at UCSD Health between January 1, 2014, and September 30, 2016.
They excluded units given to patients with gastrointestinal bleeding and units given within 12 hours of a surgical procedure. The team said they excluded these transfusions because of the higher probability of unstable blood volume or special circumstances that might justify off-protocol transfusion.
The researchers then calculated the rate of inpatient RBC units transfused per 1000 patient-days (without exclusions), the percentage of inpatient RBC units transfused for patients with Hb ≥ 7 g/dL, and the percentage of multi-unit RBC transfusions, divided into 3 periods:
- Pre-intervention or baseline (January 1, 2014–September 30, 2014)
- During the intervention (October 1, 2014–April 30, 2015)
- Post-intervention (May 1, 2015–September 30, 2016).
There were 36,386 RBC units administered during the study period, which was 464,424 patient days.
Multi-unit transfusions decreased from 59.9% at baseline to 41.7% during the intervention period and 19.7% during the post-intervention period (P<0.0001).
Transfusions in patients with Hb values ≥ 7 g/dL decreased from 72.3% at baseline to 57.8% during the intervention period and 38.0% during the post-intervention period (P<0.0001).
The total RBC transfusion rate (units per 1000 patient-days) decreased from 89.8 at baseline to 78.1 during the intervention period and 72.7 during the post-intervention period (P<0.0001).
The estimated savings, based on a cost of $250 per RBC unit, was about $1,050,750 per year.
GEP classifier predicts risk in MM
A gene expression profiling (GEP) classifier can accurately identify high- and low-risk patients with multiple myeloma (MM), according to research published in Clinical Lymphoma, Myeloma and Leukemia.
The GEP classifier is known as SKY92, and researchers found they could accurately identify high-risk MM patients using SKY92 alone.
By combining SKY92 with the International Staging System (ISS), the team was able to identify low-risk MM patients as well.
“We have demonstrated the prognostic value of SKY92 not only as a marker of high risk, but also as a marker of low risk when combined with ISS,” said study author Erik H. van Beers, PhD, vice president of genomics at SkylineDx, the company that developed SKY92.
“Both validated markers may serve as the basis for the discovery of improved individualized therapies for patients with multiple myeloma.”
Dr van Beers and his colleagues compared 8 risk-assessment platforms to analyze gene expression data from 91 newly diagnosed MM patients. The patients were included in an independent dataset amassed by the Multiple Myeloma Research Foundation and the Multiple Myeloma Genomics Initiative (MMRF/MMGI).
The researchers used the GEP classifiers SKY92, UAMS70, UAMS80, IFM15, HM19, Cancer Testis Antigen, Centrosome Index, and Proliferation Index to identify high-risk patients.
Of the 8 classifiers, SKY92 identified the largest proportion (21%) of high-risk cases and also attained the highest Cox proportional hazard ratio (8.2) for overall survival (OS).
Additionally, SKY92 predicted 9 of the 13 (60%) deaths that occurred at 2 years and 16 of the 31 (52%) deaths at 5 years, a predictive rate that was higher than any of the other classifiers.
Dr van Beers and his colleagues also combined the SKY92 standard-risk classifier with the ISS to identify low-risk patients in the MMRF/MMGI cohort. This combination was recently identified as a marker for detecting low-risk MM.*
The SKY92/ISS marker identified 42% of patients as low risk, with their median OS not reached at 96 months. The low-risk classification was strongly supported by the achieved hazard ratio of 10.
“The MMRF/MMGI dataset was not part of our previous discovery*, which demonstrated that the combination of SKY92 and ISS identifies the lowest-risk patients with high accuracy and leaves the smallest proportion of patients identified as intermediate risk,” Dr van Beers said. “Thus, the dataset remained available for independent validation—an important factor in the adoption of gene expression profiling tools.”
“We are pleased to have validated the SKY92/ISS low-risk marker by applying it to the well-characterized MMRF/MMGI cohort,” added study author Rafael Fonseca, MD, of the Mayo Clinic in Scottsdale, Arizona. “Our findings further strengthen the prognostic utility of the combination marker.”
*Kuiper R, van Duin M, van Vliet, MH, et al. Prediction of high- and low-risk multiple myeloma based on gene expression and the International Staging System. Blood. 2015;126(17):1996-2004.
A gene expression profiling (GEP) classifier can accurately identify high- and low-risk patients with multiple myeloma (MM), according to research published in Clinical Lymphoma, Myeloma and Leukemia.
The GEP classifier is known as SKY92, and researchers found they could accurately identify high-risk MM patients using SKY92 alone.
By combining SKY92 with the International Staging System (ISS), the team was able to identify low-risk MM patients as well.
“We have demonstrated the prognostic value of SKY92 not only as a marker of high risk, but also as a marker of low risk when combined with ISS,” said study author Erik H. van Beers, PhD, vice president of genomics at SkylineDx, the company that developed SKY92.
“Both validated markers may serve as the basis for the discovery of improved individualized therapies for patients with multiple myeloma.”
Dr van Beers and his colleagues compared 8 risk-assessment platforms to analyze gene expression data from 91 newly diagnosed MM patients. The patients were included in an independent dataset amassed by the Multiple Myeloma Research Foundation and the Multiple Myeloma Genomics Initiative (MMRF/MMGI).
The researchers used the GEP classifiers SKY92, UAMS70, UAMS80, IFM15, HM19, Cancer Testis Antigen, Centrosome Index, and Proliferation Index to identify high-risk patients.
Of the 8 classifiers, SKY92 identified the largest proportion (21%) of high-risk cases and also attained the highest Cox proportional hazard ratio (8.2) for overall survival (OS).
Additionally, SKY92 predicted 9 of the 13 (60%) deaths that occurred at 2 years and 16 of the 31 (52%) deaths at 5 years, a predictive rate that was higher than any of the other classifiers.
Dr van Beers and his colleagues also combined the SKY92 standard-risk classifier with the ISS to identify low-risk patients in the MMRF/MMGI cohort. This combination was recently identified as a marker for detecting low-risk MM.*
The SKY92/ISS marker identified 42% of patients as low risk, with their median OS not reached at 96 months. The low-risk classification was strongly supported by the achieved hazard ratio of 10.
“The MMRF/MMGI dataset was not part of our previous discovery*, which demonstrated that the combination of SKY92 and ISS identifies the lowest-risk patients with high accuracy and leaves the smallest proportion of patients identified as intermediate risk,” Dr van Beers said. “Thus, the dataset remained available for independent validation—an important factor in the adoption of gene expression profiling tools.”
“We are pleased to have validated the SKY92/ISS low-risk marker by applying it to the well-characterized MMRF/MMGI cohort,” added study author Rafael Fonseca, MD, of the Mayo Clinic in Scottsdale, Arizona. “Our findings further strengthen the prognostic utility of the combination marker.”
*Kuiper R, van Duin M, van Vliet, MH, et al. Prediction of high- and low-risk multiple myeloma based on gene expression and the International Staging System. Blood. 2015;126(17):1996-2004.
A gene expression profiling (GEP) classifier can accurately identify high- and low-risk patients with multiple myeloma (MM), according to research published in Clinical Lymphoma, Myeloma and Leukemia.
The GEP classifier is known as SKY92, and researchers found they could accurately identify high-risk MM patients using SKY92 alone.
By combining SKY92 with the International Staging System (ISS), the team was able to identify low-risk MM patients as well.
“We have demonstrated the prognostic value of SKY92 not only as a marker of high risk, but also as a marker of low risk when combined with ISS,” said study author Erik H. van Beers, PhD, vice president of genomics at SkylineDx, the company that developed SKY92.
“Both validated markers may serve as the basis for the discovery of improved individualized therapies for patients with multiple myeloma.”
Dr van Beers and his colleagues compared 8 risk-assessment platforms to analyze gene expression data from 91 newly diagnosed MM patients. The patients were included in an independent dataset amassed by the Multiple Myeloma Research Foundation and the Multiple Myeloma Genomics Initiative (MMRF/MMGI).
The researchers used the GEP classifiers SKY92, UAMS70, UAMS80, IFM15, HM19, Cancer Testis Antigen, Centrosome Index, and Proliferation Index to identify high-risk patients.
Of the 8 classifiers, SKY92 identified the largest proportion (21%) of high-risk cases and also attained the highest Cox proportional hazard ratio (8.2) for overall survival (OS).
Additionally, SKY92 predicted 9 of the 13 (60%) deaths that occurred at 2 years and 16 of the 31 (52%) deaths at 5 years, a predictive rate that was higher than any of the other classifiers.
Dr van Beers and his colleagues also combined the SKY92 standard-risk classifier with the ISS to identify low-risk patients in the MMRF/MMGI cohort. This combination was recently identified as a marker for detecting low-risk MM.*
The SKY92/ISS marker identified 42% of patients as low risk, with their median OS not reached at 96 months. The low-risk classification was strongly supported by the achieved hazard ratio of 10.
“The MMRF/MMGI dataset was not part of our previous discovery*, which demonstrated that the combination of SKY92 and ISS identifies the lowest-risk patients with high accuracy and leaves the smallest proportion of patients identified as intermediate risk,” Dr van Beers said. “Thus, the dataset remained available for independent validation—an important factor in the adoption of gene expression profiling tools.”
“We are pleased to have validated the SKY92/ISS low-risk marker by applying it to the well-characterized MMRF/MMGI cohort,” added study author Rafael Fonseca, MD, of the Mayo Clinic in Scottsdale, Arizona. “Our findings further strengthen the prognostic utility of the combination marker.”
*Kuiper R, van Duin M, van Vliet, MH, et al. Prediction of high- and low-risk multiple myeloma based on gene expression and the International Staging System. Blood. 2015;126(17):1996-2004.
Product granted fast track designation for aTTP
The US Food and Drug Administration (FDA) has granted fast track designation to caplacizumab, an anti-von Willebrand factor (vWF) nanobody being developed for the treatment of acquired thrombotic thrombocytopenic purpura (aTTP).
The FDA’s fast track program is designed to facilitate the development and expedite the review of products intended to treat or prevent serious or life-threatening conditions and address unmet medical need.
Through the fast track program, a product may be eligible for priority review. In addition, the company developing the product may be allowed to submit sections of the new drug application or biologics license application on a rolling basis as data become available.
Fast track designation also provides the company with opportunities for more frequent meetings and written communications with the FDA.
About caplacizumab
Caplacizumab is a bivalent anti-vWF nanobody being developed by Ablynx. Caplacizumab works by blocking the interaction of ultra-large vWF multimers with platelets, having an immediate effect on platelet aggregation and the ensuing formation and accumulation of the micro-clots that cause the severe thrombocytopenia, tissue ischemia, and organ dysfunction that occurs in patients with aTTP.
Researchers evaluated the efficacy and safety of caplacizumab, given with standard care for aTTP, in the phase 2 TITAN trial.
The trial enrolled 75 patients with aTTP. They all received the standard of care—daily plasma exchange and immunosuppressive therapy. Thirty-six patients were randomized to receive caplacizumab as well, and 39 were randomized to placebo.
The study’s primary endpoint was time to response (platelet count normalization). Patients in the caplacizumab arm had a 39% reduction in the median time to response compared to patients in the placebo arm (P=0.005).
The rate of confirmed response was 86.1% (n=31) in the caplacizumab arm and 71.8% (n=28) in the placebo arm.
There were more relapses in the caplacizumab arm than the placebo arm—8 (22.2%) and 0, respectively. Relapse was defined as a TTP event occurring more than 30 days after the end of daily plasma exchange.
There were fewer exacerbations in the caplacizumab arm than the placebo arm—3 (8.3%) and 11 (28.2%), respectively. Exacerbation was defined as recurrent thrombocytopenia within 30 days of the end of daily plasma exchange that required re-initiation of daily exchange.
The rate of adverse events thought to be related to the study drug was 17% in the caplacizumab arm and 11% in the placebo arm. The rate of events that were possibly related was 54% and 8%, respectively.
A lower proportion of subjects in the caplacizumab arm experienced one or more major thromboembolic events or died, compared to the placebo arm—11.4% and 43.2%, respectively.
In addition, fewer caplacizumab-treated patients were refractory to treatment—5.7% vs 21.6%.
There were 2 deaths in the placebo arm, and both of those patients were refractory to treatment. There were no deaths reported in the caplacizumab arm.
Now, researchers are evaluating caplacizumab in the phase 3 HERCULES trial (NCT02553317). Results from this study are anticipated in the second half of 2017 are expected to support a planned biologics license application filing in the US in 2018.
The US Food and Drug Administration (FDA) has granted fast track designation to caplacizumab, an anti-von Willebrand factor (vWF) nanobody being developed for the treatment of acquired thrombotic thrombocytopenic purpura (aTTP).
The FDA’s fast track program is designed to facilitate the development and expedite the review of products intended to treat or prevent serious or life-threatening conditions and address unmet medical need.
Through the fast track program, a product may be eligible for priority review. In addition, the company developing the product may be allowed to submit sections of the new drug application or biologics license application on a rolling basis as data become available.
Fast track designation also provides the company with opportunities for more frequent meetings and written communications with the FDA.
About caplacizumab
Caplacizumab is a bivalent anti-vWF nanobody being developed by Ablynx. Caplacizumab works by blocking the interaction of ultra-large vWF multimers with platelets, having an immediate effect on platelet aggregation and the ensuing formation and accumulation of the micro-clots that cause the severe thrombocytopenia, tissue ischemia, and organ dysfunction that occurs in patients with aTTP.
Researchers evaluated the efficacy and safety of caplacizumab, given with standard care for aTTP, in the phase 2 TITAN trial.
The trial enrolled 75 patients with aTTP. They all received the standard of care—daily plasma exchange and immunosuppressive therapy. Thirty-six patients were randomized to receive caplacizumab as well, and 39 were randomized to placebo.
The study’s primary endpoint was time to response (platelet count normalization). Patients in the caplacizumab arm had a 39% reduction in the median time to response compared to patients in the placebo arm (P=0.005).
The rate of confirmed response was 86.1% (n=31) in the caplacizumab arm and 71.8% (n=28) in the placebo arm.
There were more relapses in the caplacizumab arm than the placebo arm—8 (22.2%) and 0, respectively. Relapse was defined as a TTP event occurring more than 30 days after the end of daily plasma exchange.
There were fewer exacerbations in the caplacizumab arm than the placebo arm—3 (8.3%) and 11 (28.2%), respectively. Exacerbation was defined as recurrent thrombocytopenia within 30 days of the end of daily plasma exchange that required re-initiation of daily exchange.
The rate of adverse events thought to be related to the study drug was 17% in the caplacizumab arm and 11% in the placebo arm. The rate of events that were possibly related was 54% and 8%, respectively.
A lower proportion of subjects in the caplacizumab arm experienced one or more major thromboembolic events or died, compared to the placebo arm—11.4% and 43.2%, respectively.
In addition, fewer caplacizumab-treated patients were refractory to treatment—5.7% vs 21.6%.
There were 2 deaths in the placebo arm, and both of those patients were refractory to treatment. There were no deaths reported in the caplacizumab arm.
Now, researchers are evaluating caplacizumab in the phase 3 HERCULES trial (NCT02553317). Results from this study are anticipated in the second half of 2017 are expected to support a planned biologics license application filing in the US in 2018.
The US Food and Drug Administration (FDA) has granted fast track designation to caplacizumab, an anti-von Willebrand factor (vWF) nanobody being developed for the treatment of acquired thrombotic thrombocytopenic purpura (aTTP).
The FDA’s fast track program is designed to facilitate the development and expedite the review of products intended to treat or prevent serious or life-threatening conditions and address unmet medical need.
Through the fast track program, a product may be eligible for priority review. In addition, the company developing the product may be allowed to submit sections of the new drug application or biologics license application on a rolling basis as data become available.
Fast track designation also provides the company with opportunities for more frequent meetings and written communications with the FDA.
About caplacizumab
Caplacizumab is a bivalent anti-vWF nanobody being developed by Ablynx. Caplacizumab works by blocking the interaction of ultra-large vWF multimers with platelets, having an immediate effect on platelet aggregation and the ensuing formation and accumulation of the micro-clots that cause the severe thrombocytopenia, tissue ischemia, and organ dysfunction that occurs in patients with aTTP.
Researchers evaluated the efficacy and safety of caplacizumab, given with standard care for aTTP, in the phase 2 TITAN trial.
The trial enrolled 75 patients with aTTP. They all received the standard of care—daily plasma exchange and immunosuppressive therapy. Thirty-six patients were randomized to receive caplacizumab as well, and 39 were randomized to placebo.
The study’s primary endpoint was time to response (platelet count normalization). Patients in the caplacizumab arm had a 39% reduction in the median time to response compared to patients in the placebo arm (P=0.005).
The rate of confirmed response was 86.1% (n=31) in the caplacizumab arm and 71.8% (n=28) in the placebo arm.
There were more relapses in the caplacizumab arm than the placebo arm—8 (22.2%) and 0, respectively. Relapse was defined as a TTP event occurring more than 30 days after the end of daily plasma exchange.
There were fewer exacerbations in the caplacizumab arm than the placebo arm—3 (8.3%) and 11 (28.2%), respectively. Exacerbation was defined as recurrent thrombocytopenia within 30 days of the end of daily plasma exchange that required re-initiation of daily exchange.
The rate of adverse events thought to be related to the study drug was 17% in the caplacizumab arm and 11% in the placebo arm. The rate of events that were possibly related was 54% and 8%, respectively.
A lower proportion of subjects in the caplacizumab arm experienced one or more major thromboembolic events or died, compared to the placebo arm—11.4% and 43.2%, respectively.
In addition, fewer caplacizumab-treated patients were refractory to treatment—5.7% vs 21.6%.
There were 2 deaths in the placebo arm, and both of those patients were refractory to treatment. There were no deaths reported in the caplacizumab arm.
Now, researchers are evaluating caplacizumab in the phase 3 HERCULES trial (NCT02553317). Results from this study are anticipated in the second half of 2017 are expected to support a planned biologics license application filing in the US in 2018.