User login
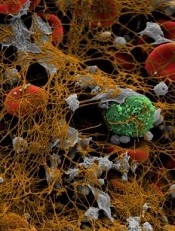
Authority on hematologic malignancies dies
Physician, researcher, and educator H. Jean Khoury, MD, recently passed away.
He died on Monday, May 22, at the age of 50, after a year-long battle with esophageal cancer.
Dr Khoury led the division of hematology at Winship Cancer Institute of Emory University in Atlanta, Georgia.
He was considered an authority on hematologic malignancies, particularly chronic myeloid leukemia (CML), acute leukemia, and myelodysplastic syndromes (MDS).
Dr Khoury joined Winship Cancer Institute in 2004 as director of the Leukemia Service and associate professor in the Emory School of Medicine.
In 2009, he was promoted to professor and director of the Division of Hematology in the Department of Hematology and Medical Oncology, and he was later named to the R. Randall Rollins Chair in Oncology.
“We are all deeply grieving the loss of this remarkable man who gave so much to Winship,” said Walter J. Curran, Jr, MD, Winship Cancer Institute’s executive director.
“His enthusiasm and love for his patients and his commitment to lessening the burden of cancer for all has been unwavering throughout his life.”
A native of Beirut, Lebanon, Dr Khoury came to the Winship Cancer Institute from Washington University in St Louis, Missouri, where he served on the faculty after completing a fellowship in hematology-oncology.
He earned his medical degree from the Université Catholique de Louvain in Brussels, Belgium, and completed a residency in internal medicine at Memorial Medical Center in Savannah, Georgia.
Dr Khoury was recruited to Winship Cancer Institute by Fadlo R. Khuri, MD, former deputy director of the institute and now president of the American University of Beirut. What he first saw in Dr Khoury was someone who was “in the best sense, a disruptive presence.”
“What you always want in a leader is someone who’s not afraid to be wrong, to take risks,” Dr Khuri said. “Being wrong disrupts the pattern, and Jean was very brave. He didn’t like business as usual, and that showed in the way he took about redeveloping the hematology division, the leukemia program, and his interactions with the transplant division, with faculty, and all across Winship.”
According to his colleagues, Dr Khoury’s guiding principle was how to improve his patients’ lives, whether through research discoveries or through compassionate care.
Even after being diagnosed with cancer himself, Dr Khoury continued to see patients and carry on his work in the clinic and his research.
Dr Khoury pioneered the development of personalized treatment for CML patients and better approaches to improve quality of life for survivors. His research focused on drug development in leukemia and MDS, genomic abnormalities in leukemia, development of cost-effective practice models, and outcome analysis of bone marrow transplant.
He conducted several leukemia and transplant clinical trials, including trials that led to the approval of drugs such as imatinib, dasatinib, and nilotinib.
Dr Khoury received the Georgia Cancer Coalition Distinguished Cancer Scholarship, which allowed for establishment of the Hematological Disorders Tissue Bank at Emory, which now contains annotated germline and somatic samples from more than 800 patients with various hematologic disorders.
Dr Khoury died at home with his family by his side. He is survived by his wife, Angela, and 3 children, Mikhail, Iman, and Alya.
In lieu of flowers, the family requests that contributions be made to a new fund at Winship Cancer Institute that will memorialize the life and work of Dr Khoury by supporting a fellowship program that was so meaningful to him.
Contributions, marked in Memory of Dr H. Jean Khoury, can be sent to Winship Cancer Institute of Emory University, Office of Gift Records, Emory University, 1762 Clifton Rd. NE, Suite 1400, Atlanta, GA 30322. Gifts can also be made online.
There will be a memorial service for Dr Khoury on Wednesday, May 31, at 4:30 pm at Glenn Memorial Church, 1652 North Decatur Road in Atlanta, Georgia. ![]()
Physician, researcher, and educator H. Jean Khoury, MD, recently passed away.
He died on Monday, May 22, at the age of 50, after a year-long battle with esophageal cancer.
Dr Khoury led the division of hematology at Winship Cancer Institute of Emory University in Atlanta, Georgia.
He was considered an authority on hematologic malignancies, particularly chronic myeloid leukemia (CML), acute leukemia, and myelodysplastic syndromes (MDS).
Dr Khoury joined Winship Cancer Institute in 2004 as director of the Leukemia Service and associate professor in the Emory School of Medicine.
In 2009, he was promoted to professor and director of the Division of Hematology in the Department of Hematology and Medical Oncology, and he was later named to the R. Randall Rollins Chair in Oncology.
“We are all deeply grieving the loss of this remarkable man who gave so much to Winship,” said Walter J. Curran, Jr, MD, Winship Cancer Institute’s executive director.
“His enthusiasm and love for his patients and his commitment to lessening the burden of cancer for all has been unwavering throughout his life.”
A native of Beirut, Lebanon, Dr Khoury came to the Winship Cancer Institute from Washington University in St Louis, Missouri, where he served on the faculty after completing a fellowship in hematology-oncology.
He earned his medical degree from the Université Catholique de Louvain in Brussels, Belgium, and completed a residency in internal medicine at Memorial Medical Center in Savannah, Georgia.
Dr Khoury was recruited to Winship Cancer Institute by Fadlo R. Khuri, MD, former deputy director of the institute and now president of the American University of Beirut. What he first saw in Dr Khoury was someone who was “in the best sense, a disruptive presence.”
“What you always want in a leader is someone who’s not afraid to be wrong, to take risks,” Dr Khuri said. “Being wrong disrupts the pattern, and Jean was very brave. He didn’t like business as usual, and that showed in the way he took about redeveloping the hematology division, the leukemia program, and his interactions with the transplant division, with faculty, and all across Winship.”
According to his colleagues, Dr Khoury’s guiding principle was how to improve his patients’ lives, whether through research discoveries or through compassionate care.
Even after being diagnosed with cancer himself, Dr Khoury continued to see patients and carry on his work in the clinic and his research.
Dr Khoury pioneered the development of personalized treatment for CML patients and better approaches to improve quality of life for survivors. His research focused on drug development in leukemia and MDS, genomic abnormalities in leukemia, development of cost-effective practice models, and outcome analysis of bone marrow transplant.
He conducted several leukemia and transplant clinical trials, including trials that led to the approval of drugs such as imatinib, dasatinib, and nilotinib.
Dr Khoury received the Georgia Cancer Coalition Distinguished Cancer Scholarship, which allowed for establishment of the Hematological Disorders Tissue Bank at Emory, which now contains annotated germline and somatic samples from more than 800 patients with various hematologic disorders.
Dr Khoury died at home with his family by his side. He is survived by his wife, Angela, and 3 children, Mikhail, Iman, and Alya.
In lieu of flowers, the family requests that contributions be made to a new fund at Winship Cancer Institute that will memorialize the life and work of Dr Khoury by supporting a fellowship program that was so meaningful to him.
Contributions, marked in Memory of Dr H. Jean Khoury, can be sent to Winship Cancer Institute of Emory University, Office of Gift Records, Emory University, 1762 Clifton Rd. NE, Suite 1400, Atlanta, GA 30322. Gifts can also be made online.
There will be a memorial service for Dr Khoury on Wednesday, May 31, at 4:30 pm at Glenn Memorial Church, 1652 North Decatur Road in Atlanta, Georgia. ![]()
Physician, researcher, and educator H. Jean Khoury, MD, recently passed away.
He died on Monday, May 22, at the age of 50, after a year-long battle with esophageal cancer.
Dr Khoury led the division of hematology at Winship Cancer Institute of Emory University in Atlanta, Georgia.
He was considered an authority on hematologic malignancies, particularly chronic myeloid leukemia (CML), acute leukemia, and myelodysplastic syndromes (MDS).
Dr Khoury joined Winship Cancer Institute in 2004 as director of the Leukemia Service and associate professor in the Emory School of Medicine.
In 2009, he was promoted to professor and director of the Division of Hematology in the Department of Hematology and Medical Oncology, and he was later named to the R. Randall Rollins Chair in Oncology.
“We are all deeply grieving the loss of this remarkable man who gave so much to Winship,” said Walter J. Curran, Jr, MD, Winship Cancer Institute’s executive director.
“His enthusiasm and love for his patients and his commitment to lessening the burden of cancer for all has been unwavering throughout his life.”
A native of Beirut, Lebanon, Dr Khoury came to the Winship Cancer Institute from Washington University in St Louis, Missouri, where he served on the faculty after completing a fellowship in hematology-oncology.
He earned his medical degree from the Université Catholique de Louvain in Brussels, Belgium, and completed a residency in internal medicine at Memorial Medical Center in Savannah, Georgia.
Dr Khoury was recruited to Winship Cancer Institute by Fadlo R. Khuri, MD, former deputy director of the institute and now president of the American University of Beirut. What he first saw in Dr Khoury was someone who was “in the best sense, a disruptive presence.”
“What you always want in a leader is someone who’s not afraid to be wrong, to take risks,” Dr Khuri said. “Being wrong disrupts the pattern, and Jean was very brave. He didn’t like business as usual, and that showed in the way he took about redeveloping the hematology division, the leukemia program, and his interactions with the transplant division, with faculty, and all across Winship.”
According to his colleagues, Dr Khoury’s guiding principle was how to improve his patients’ lives, whether through research discoveries or through compassionate care.
Even after being diagnosed with cancer himself, Dr Khoury continued to see patients and carry on his work in the clinic and his research.
Dr Khoury pioneered the development of personalized treatment for CML patients and better approaches to improve quality of life for survivors. His research focused on drug development in leukemia and MDS, genomic abnormalities in leukemia, development of cost-effective practice models, and outcome analysis of bone marrow transplant.
He conducted several leukemia and transplant clinical trials, including trials that led to the approval of drugs such as imatinib, dasatinib, and nilotinib.
Dr Khoury received the Georgia Cancer Coalition Distinguished Cancer Scholarship, which allowed for establishment of the Hematological Disorders Tissue Bank at Emory, which now contains annotated germline and somatic samples from more than 800 patients with various hematologic disorders.
Dr Khoury died at home with his family by his side. He is survived by his wife, Angela, and 3 children, Mikhail, Iman, and Alya.
In lieu of flowers, the family requests that contributions be made to a new fund at Winship Cancer Institute that will memorialize the life and work of Dr Khoury by supporting a fellowship program that was so meaningful to him.
Contributions, marked in Memory of Dr H. Jean Khoury, can be sent to Winship Cancer Institute of Emory University, Office of Gift Records, Emory University, 1762 Clifton Rd. NE, Suite 1400, Atlanta, GA 30322. Gifts can also be made online.
There will be a memorial service for Dr Khoury on Wednesday, May 31, at 4:30 pm at Glenn Memorial Church, 1652 North Decatur Road in Atlanta, Georgia. ![]()
Group uses mobile lab to track spread of Zika in Brazil
Researchers say they have uncovered the origin of Zika virus in Brazil and tracked the spread of the virus across the country.
The team traveled across northeast Brazil in a mobile genomics lab, analyzing samples from Zika patients.
The researchers found that Zika’s establishment within Brazil—and its spread from there to other regions—occurred before Zika transmission in the Americas was first discovered.
The team reported their findings in Nature.
To gain insight into the epidemiology and evolution of Zika virus, the researchers traveled 2000 km across northeast Brazil in June of last year. The team traveled in a minibus equipped with mobile DNA sequencing capabilities and tested samples from more than 1300 patients infected with the virus.
“Despite there being probably millions of cases of the Zika virus in Brazil, there was only a handful of known virus genomes prior to our work,” said Nuno Faria, PhD, of the University of Oxford in the UK.
“A better understanding of Zika virus genetic diversity is critical to vaccine design and also to identify areas where surveillance is most needed.”
“We generated Zika virus genomes to establish the virus epidemic history in the Americas,” said Oliver Pybus, DPhil, also of the University of Oxford.
“We showed that the virus was present in Brazil for a full year prior to the first confirmed cases in May 2015. We also found that northeast Brazil, which was the region with the most recorded cases of Zika and microcephaly, was the nexus of the epidemic in Brazil and played a key role in its spread within Brazil to major urban centers, such as Rio de Janeiro and São Paulo, before spreading across the Americas. We now have a better understanding of the epidemiology of the virus.”
During the genome sequencing journey across Brazil, the researchers used the portable MinION DNA sequencer from the company Oxford Nanopore Technologies, which started as an Oxford University spinout company. The device weighs less than 100 g and is powered by the USB port of a laptop.
“Genome sequencing has become a powerful tool for studying emerging infectious diseases,” said Nick Loman, PhD, of the University of Birmingham in the UK.
“However, genome sequencing directly from clinical samples without isolation remains challenging for viruses such as Zika. We developed a new protocol that allows for real-time genomic sequencing—something of vital importance when managing viral outbreaks, as it can provide real insight into how a virus is spreading, transmitting and evolving.”
“What’s more, using equipment like portable nanopore sequencing, we were able to carry out emergency epidemiological research more quickly, to get immediate results when working in the field, as on the road in Brazil. This new protocol will doubtless prove hugely beneficial to researchers working in remote areas around the world during times of viral outbreaks.” ![]()
Researchers say they have uncovered the origin of Zika virus in Brazil and tracked the spread of the virus across the country.
The team traveled across northeast Brazil in a mobile genomics lab, analyzing samples from Zika patients.
The researchers found that Zika’s establishment within Brazil—and its spread from there to other regions—occurred before Zika transmission in the Americas was first discovered.
The team reported their findings in Nature.
To gain insight into the epidemiology and evolution of Zika virus, the researchers traveled 2000 km across northeast Brazil in June of last year. The team traveled in a minibus equipped with mobile DNA sequencing capabilities and tested samples from more than 1300 patients infected with the virus.
“Despite there being probably millions of cases of the Zika virus in Brazil, there was only a handful of known virus genomes prior to our work,” said Nuno Faria, PhD, of the University of Oxford in the UK.
“A better understanding of Zika virus genetic diversity is critical to vaccine design and also to identify areas where surveillance is most needed.”
“We generated Zika virus genomes to establish the virus epidemic history in the Americas,” said Oliver Pybus, DPhil, also of the University of Oxford.
“We showed that the virus was present in Brazil for a full year prior to the first confirmed cases in May 2015. We also found that northeast Brazil, which was the region with the most recorded cases of Zika and microcephaly, was the nexus of the epidemic in Brazil and played a key role in its spread within Brazil to major urban centers, such as Rio de Janeiro and São Paulo, before spreading across the Americas. We now have a better understanding of the epidemiology of the virus.”
During the genome sequencing journey across Brazil, the researchers used the portable MinION DNA sequencer from the company Oxford Nanopore Technologies, which started as an Oxford University spinout company. The device weighs less than 100 g and is powered by the USB port of a laptop.
“Genome sequencing has become a powerful tool for studying emerging infectious diseases,” said Nick Loman, PhD, of the University of Birmingham in the UK.
“However, genome sequencing directly from clinical samples without isolation remains challenging for viruses such as Zika. We developed a new protocol that allows for real-time genomic sequencing—something of vital importance when managing viral outbreaks, as it can provide real insight into how a virus is spreading, transmitting and evolving.”
“What’s more, using equipment like portable nanopore sequencing, we were able to carry out emergency epidemiological research more quickly, to get immediate results when working in the field, as on the road in Brazil. This new protocol will doubtless prove hugely beneficial to researchers working in remote areas around the world during times of viral outbreaks.” ![]()
Researchers say they have uncovered the origin of Zika virus in Brazil and tracked the spread of the virus across the country.
The team traveled across northeast Brazil in a mobile genomics lab, analyzing samples from Zika patients.
The researchers found that Zika’s establishment within Brazil—and its spread from there to other regions—occurred before Zika transmission in the Americas was first discovered.
The team reported their findings in Nature.
To gain insight into the epidemiology and evolution of Zika virus, the researchers traveled 2000 km across northeast Brazil in June of last year. The team traveled in a minibus equipped with mobile DNA sequencing capabilities and tested samples from more than 1300 patients infected with the virus.
“Despite there being probably millions of cases of the Zika virus in Brazil, there was only a handful of known virus genomes prior to our work,” said Nuno Faria, PhD, of the University of Oxford in the UK.
“A better understanding of Zika virus genetic diversity is critical to vaccine design and also to identify areas where surveillance is most needed.”
“We generated Zika virus genomes to establish the virus epidemic history in the Americas,” said Oliver Pybus, DPhil, also of the University of Oxford.
“We showed that the virus was present in Brazil for a full year prior to the first confirmed cases in May 2015. We also found that northeast Brazil, which was the region with the most recorded cases of Zika and microcephaly, was the nexus of the epidemic in Brazil and played a key role in its spread within Brazil to major urban centers, such as Rio de Janeiro and São Paulo, before spreading across the Americas. We now have a better understanding of the epidemiology of the virus.”
During the genome sequencing journey across Brazil, the researchers used the portable MinION DNA sequencer from the company Oxford Nanopore Technologies, which started as an Oxford University spinout company. The device weighs less than 100 g and is powered by the USB port of a laptop.
“Genome sequencing has become a powerful tool for studying emerging infectious diseases,” said Nick Loman, PhD, of the University of Birmingham in the UK.
“However, genome sequencing directly from clinical samples without isolation remains challenging for viruses such as Zika. We developed a new protocol that allows for real-time genomic sequencing—something of vital importance when managing viral outbreaks, as it can provide real insight into how a virus is spreading, transmitting and evolving.”
“What’s more, using equipment like portable nanopore sequencing, we were able to carry out emergency epidemiological research more quickly, to get immediate results when working in the field, as on the road in Brazil. This new protocol will doubtless prove hugely beneficial to researchers working in remote areas around the world during times of viral outbreaks.” ![]()
Zika virus spread undetected in the Americas, team says
New research suggests the Zika virus spread quickly in the Americas and then diverged into distinct genetic groups.
Researchers performed genetic analysis on samples collected as the virus spread throughout the Americas after its introduction in 2013 or 2014.
The team found that Zika circulated undetected for up to a year in some regions before it came to the attention of public health authorities.
Genetic sequencing also enabled the researchers to recreate the epidemiological and evolutionary paths the virus took as it spread and split into the distinct subtypes—or clades—that have been detected in the Americas.
Hayden C. Metsky, a PhD student at the Broad Institute of MIT and Harvard, Cambridge, Massachusetts, and his colleagues reported these findings in Nature.
The researchers reconstructed Zika’s dispersal by sequencing genetic material collected from hundreds of patients in 10 countries and territories.
The team eventually amassed a database of 110 complete or partial Zika virus genomes—the largest collection to date—which they analyzed along with 64 published and publicly shared genomes.
Based on changes to the viral genome that accumulated as the disease moved through new populations, the researchers concluded that Zika virus spread rapidly upon its initial introduction in Brazil, likely sometime in 2013.
Later, at several points in early to mid-2015, the virus separated into at least 3 clades—distinct genetic groups whose members share a common ancestor—in Colombia, Honduras, and Puerto Rico, as well as a fourth type found in parts of the Caribbean and the continental US.
The researchers believe these findings could have a direct impact on public health, informing disease surveillance and the development of diagnostic tests. ![]()
New research suggests the Zika virus spread quickly in the Americas and then diverged into distinct genetic groups.
Researchers performed genetic analysis on samples collected as the virus spread throughout the Americas after its introduction in 2013 or 2014.
The team found that Zika circulated undetected for up to a year in some regions before it came to the attention of public health authorities.
Genetic sequencing also enabled the researchers to recreate the epidemiological and evolutionary paths the virus took as it spread and split into the distinct subtypes—or clades—that have been detected in the Americas.
Hayden C. Metsky, a PhD student at the Broad Institute of MIT and Harvard, Cambridge, Massachusetts, and his colleagues reported these findings in Nature.
The researchers reconstructed Zika’s dispersal by sequencing genetic material collected from hundreds of patients in 10 countries and territories.
The team eventually amassed a database of 110 complete or partial Zika virus genomes—the largest collection to date—which they analyzed along with 64 published and publicly shared genomes.
Based on changes to the viral genome that accumulated as the disease moved through new populations, the researchers concluded that Zika virus spread rapidly upon its initial introduction in Brazil, likely sometime in 2013.
Later, at several points in early to mid-2015, the virus separated into at least 3 clades—distinct genetic groups whose members share a common ancestor—in Colombia, Honduras, and Puerto Rico, as well as a fourth type found in parts of the Caribbean and the continental US.
The researchers believe these findings could have a direct impact on public health, informing disease surveillance and the development of diagnostic tests. ![]()
New research suggests the Zika virus spread quickly in the Americas and then diverged into distinct genetic groups.
Researchers performed genetic analysis on samples collected as the virus spread throughout the Americas after its introduction in 2013 or 2014.
The team found that Zika circulated undetected for up to a year in some regions before it came to the attention of public health authorities.
Genetic sequencing also enabled the researchers to recreate the epidemiological and evolutionary paths the virus took as it spread and split into the distinct subtypes—or clades—that have been detected in the Americas.
Hayden C. Metsky, a PhD student at the Broad Institute of MIT and Harvard, Cambridge, Massachusetts, and his colleagues reported these findings in Nature.
The researchers reconstructed Zika’s dispersal by sequencing genetic material collected from hundreds of patients in 10 countries and territories.
The team eventually amassed a database of 110 complete or partial Zika virus genomes—the largest collection to date—which they analyzed along with 64 published and publicly shared genomes.
Based on changes to the viral genome that accumulated as the disease moved through new populations, the researchers concluded that Zika virus spread rapidly upon its initial introduction in Brazil, likely sometime in 2013.
Later, at several points in early to mid-2015, the virus separated into at least 3 clades—distinct genetic groups whose members share a common ancestor—in Colombia, Honduras, and Puerto Rico, as well as a fourth type found in parts of the Caribbean and the continental US.
The researchers believe these findings could have a direct impact on public health, informing disease surveillance and the development of diagnostic tests. ![]()
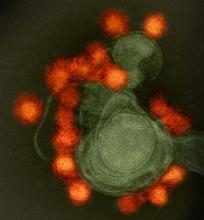
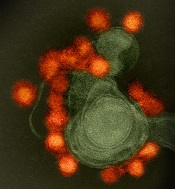
BLA for CAR T-cell therapy granted priority review
The US Food and Drug Administration (FDA) has accepted for priority review the biologics license application (BLA) for axicabtagene ciloleucel (formerly KTE-C19), a chimeric antigen receptor (CAR) T-cell therapy.
Kite Pharma, Inc. is seeking approval for axicabtagene ciloleucel as a treatment for patients with relapsed or refractory, aggressive non-Hodgkin lymphoma (NHL) who are ineligible for autologous stem cell transplant.
The FDA grants priority review to applications for products that may provide significant improvements in the treatment, diagnosis, or prevention of serious conditions.
The agency’s goal is to take action on a priority review application within 6 months of receiving it, rather than the standard 10 months.
The FDA has set a review deadline of November 29, 2017, for the axicabtagene ciloleucel BLA.
Axicabtagene ciloleucel also has breakthrough therapy designation from the FDA as a treatment for diffuse large B-cell lymphoma, transformed follicular lymphoma, and primary mediastinal B-cell lymphoma.
ZUMA-1 trial
The BLA for axicabtagene ciloleucel is supported by data from the phase 2 ZUMA-1 trial, which enrolled 111 patients with relapsed/refractory B-cell NHL.
After a single infusion of axicabtagene ciloleucel, the objective response rate was 82%. At a median follow-up of 8.7 months, 44% of patients were still in response, which included 39% of patients in complete response.
The most common grade 3 or higher adverse events were anemia (43%), neutropenia (39%), decreased neutrophil count (32%), febrile neutropenia (31%), decreased white blood cell count (29%), thrombocytopenia (24%), encephalopathy (21%), and decreased lymphocyte count (20%).
There were 3 deaths during the trial that were not due to disease progression. Two of these deaths were deemed related to axicabtagene ciloleucel. ![]()
The US Food and Drug Administration (FDA) has accepted for priority review the biologics license application (BLA) for axicabtagene ciloleucel (formerly KTE-C19), a chimeric antigen receptor (CAR) T-cell therapy.
Kite Pharma, Inc. is seeking approval for axicabtagene ciloleucel as a treatment for patients with relapsed or refractory, aggressive non-Hodgkin lymphoma (NHL) who are ineligible for autologous stem cell transplant.
The FDA grants priority review to applications for products that may provide significant improvements in the treatment, diagnosis, or prevention of serious conditions.
The agency’s goal is to take action on a priority review application within 6 months of receiving it, rather than the standard 10 months.
The FDA has set a review deadline of November 29, 2017, for the axicabtagene ciloleucel BLA.
Axicabtagene ciloleucel also has breakthrough therapy designation from the FDA as a treatment for diffuse large B-cell lymphoma, transformed follicular lymphoma, and primary mediastinal B-cell lymphoma.
ZUMA-1 trial
The BLA for axicabtagene ciloleucel is supported by data from the phase 2 ZUMA-1 trial, which enrolled 111 patients with relapsed/refractory B-cell NHL.
After a single infusion of axicabtagene ciloleucel, the objective response rate was 82%. At a median follow-up of 8.7 months, 44% of patients were still in response, which included 39% of patients in complete response.
The most common grade 3 or higher adverse events were anemia (43%), neutropenia (39%), decreased neutrophil count (32%), febrile neutropenia (31%), decreased white blood cell count (29%), thrombocytopenia (24%), encephalopathy (21%), and decreased lymphocyte count (20%).
There were 3 deaths during the trial that were not due to disease progression. Two of these deaths were deemed related to axicabtagene ciloleucel. ![]()
The US Food and Drug Administration (FDA) has accepted for priority review the biologics license application (BLA) for axicabtagene ciloleucel (formerly KTE-C19), a chimeric antigen receptor (CAR) T-cell therapy.
Kite Pharma, Inc. is seeking approval for axicabtagene ciloleucel as a treatment for patients with relapsed or refractory, aggressive non-Hodgkin lymphoma (NHL) who are ineligible for autologous stem cell transplant.
The FDA grants priority review to applications for products that may provide significant improvements in the treatment, diagnosis, or prevention of serious conditions.
The agency’s goal is to take action on a priority review application within 6 months of receiving it, rather than the standard 10 months.
The FDA has set a review deadline of November 29, 2017, for the axicabtagene ciloleucel BLA.
Axicabtagene ciloleucel also has breakthrough therapy designation from the FDA as a treatment for diffuse large B-cell lymphoma, transformed follicular lymphoma, and primary mediastinal B-cell lymphoma.
ZUMA-1 trial
The BLA for axicabtagene ciloleucel is supported by data from the phase 2 ZUMA-1 trial, which enrolled 111 patients with relapsed/refractory B-cell NHL.
After a single infusion of axicabtagene ciloleucel, the objective response rate was 82%. At a median follow-up of 8.7 months, 44% of patients were still in response, which included 39% of patients in complete response.
The most common grade 3 or higher adverse events were anemia (43%), neutropenia (39%), decreased neutrophil count (32%), febrile neutropenia (31%), decreased white blood cell count (29%), thrombocytopenia (24%), encephalopathy (21%), and decreased lymphocyte count (20%).
There were 3 deaths during the trial that were not due to disease progression. Two of these deaths were deemed related to axicabtagene ciloleucel. ![]()
How Zika arrived and spread in Florida
A study published in Nature appears to explain how Zika virus entered the US via Florida in 2016 and how the virus might re-enter the country this year.
By sequencing the Zika virus genome at different points in the outbreak, researchers created a family tree showing where cases originated and how quickly they spread.
The team discovered that transmission of Zika virus began in Florida at least 4—and potentially up to 40—times last year.
The researchers also traced most of the Zika lineages back to strains of the virus in the Caribbean.
“Without these genomes, we wouldn’t be able to reconstruct the history of how the virus moved around,” said study author Kristian G. Andersen, PhD, of The Scripps Research Institute in La Jolla, California.
“Rapid viral genome sequencing during ongoing outbreaks is a new development that has only been made possible over the last couple of years.”
Why Miami?
By sequencing Zika virus genomes from humans and mosquitoes—and analyzing travel and mosquito abundance data—the researchers found that several factors created a “perfect storm” for the spread of Zika virus in Miami.
“This study shows why Miami is special,” said study author Nathan D. Grubaugh, PhD, of The Scripps Research Institute.
First, Dr Grubaugh explained, Miami is home to year-round populations of Aedes aegypti mosquitoes, the main species that transmits Zika virus.
The area is also a significant travel hub, bringing in more international air and sea traffic than any other city in the continental US in 2016. Finally, Miami is an especially popular destination for travelers who have visited Zika-afflicted areas.
The researchers found that travel from the Caribbean islands may have significantly contributed to cases of Zika reaching Miami.
Of the 5.7 million international travelers entering Miami by flights and cruise ships between January and June of 2016, more than half arrived from the Caribbean.
Killing mosquitos produces results
The researchers believe Zika virus may have started transmission in Miami up to 40 times, but most travel-related cases did not lead to any secondary infections locally. The virus was more likely to reach a dead end than keep spreading.
The researchers found that one reason for the dead-ends was a direct connection between mosquito control efforts and disease prevention.
“We show that if you decrease the mosquito population in an area, the number of Zika infections goes down proportionally,” Dr Andersen said. “This means we can significantly limit the risk of Zika virus by focusing on mosquito control. This is not too surprising, but it’s important to show that there is an almost perfect correlation between the number of mosquitos and the number of human infections.”
Based on data from the outbreak, the researchers see potential in stopping the virus through mosquito control efforts in the US and other infected countries, instead of, for example, through travel restrictions.
“Given how many times the introductions happened, trying to restrict traffic or movement of people obviously isn’t a solution,” Dr Andersen said. “Focusing on disease prevention and mosquito control in endemic areas is likely to be a much more successful strategy.”
When the virus did spread, the researchers found that splitting Miami into designated Zika zones—often done by neighborhood or city block—didn’t accurately represent how the virus was moving.
Within each Zika zone, the researchers discovered a mixing of multiple Zika lineages, suggesting the virus wasn’t well-confined, likely moving around with infected people.
Drs Andersen and Grubaugh hope these lessons from the 2016 epidemic will help researchers and health officials respond even faster to prevent Zika’s spread in 2017.
Behind the data
Understanding Zika’s timeline required an international team of researchers and partnerships with several health agencies.
The researchers also designed a new method of genomic sequencing just to study the Zika virus. Because Zika is hard to collect in the blood of those infected, it was a challenge for the researchers to isolate enough of its genetic material for sequencing.
To solve this problem, the researchers developed 2 different protocols to break apart the genetic material they could find and reassemble it in a useful way for analysis.
With these new protocols, the researchers sequenced the virus from 28 of the reported 256 Zika cases in Florida, as well as 7 mosquito pools, to model what happened in the larger patient group.
As they worked, the researchers released their data immediately publicly to help other researchers. They hope to release more data—and analysis—in real time as cases mount in 2017.
The research was supported by the National Institutes of Health, The Pew Charitable Trusts, The Ray Thomas Foundation, a Mahan Postdoctoral Fellowship from the Computational Biology Program at Fred Hutchinson Cancer Research Center, the US Centers for Disease Control and Prevention, the European Union Seventh Framework Programme, the United States Agency for International Development Emerging Pandemic Threats Program-2 PREDICT-2, and the Defense Advanced Research Projects Agency. ![]()
A study published in Nature appears to explain how Zika virus entered the US via Florida in 2016 and how the virus might re-enter the country this year.
By sequencing the Zika virus genome at different points in the outbreak, researchers created a family tree showing where cases originated and how quickly they spread.
The team discovered that transmission of Zika virus began in Florida at least 4—and potentially up to 40—times last year.
The researchers also traced most of the Zika lineages back to strains of the virus in the Caribbean.
“Without these genomes, we wouldn’t be able to reconstruct the history of how the virus moved around,” said study author Kristian G. Andersen, PhD, of The Scripps Research Institute in La Jolla, California.
“Rapid viral genome sequencing during ongoing outbreaks is a new development that has only been made possible over the last couple of years.”
Why Miami?
By sequencing Zika virus genomes from humans and mosquitoes—and analyzing travel and mosquito abundance data—the researchers found that several factors created a “perfect storm” for the spread of Zika virus in Miami.
“This study shows why Miami is special,” said study author Nathan D. Grubaugh, PhD, of The Scripps Research Institute.
First, Dr Grubaugh explained, Miami is home to year-round populations of Aedes aegypti mosquitoes, the main species that transmits Zika virus.
The area is also a significant travel hub, bringing in more international air and sea traffic than any other city in the continental US in 2016. Finally, Miami is an especially popular destination for travelers who have visited Zika-afflicted areas.
The researchers found that travel from the Caribbean islands may have significantly contributed to cases of Zika reaching Miami.
Of the 5.7 million international travelers entering Miami by flights and cruise ships between January and June of 2016, more than half arrived from the Caribbean.
Killing mosquitos produces results
The researchers believe Zika virus may have started transmission in Miami up to 40 times, but most travel-related cases did not lead to any secondary infections locally. The virus was more likely to reach a dead end than keep spreading.
The researchers found that one reason for the dead-ends was a direct connection between mosquito control efforts and disease prevention.
“We show that if you decrease the mosquito population in an area, the number of Zika infections goes down proportionally,” Dr Andersen said. “This means we can significantly limit the risk of Zika virus by focusing on mosquito control. This is not too surprising, but it’s important to show that there is an almost perfect correlation between the number of mosquitos and the number of human infections.”
Based on data from the outbreak, the researchers see potential in stopping the virus through mosquito control efforts in the US and other infected countries, instead of, for example, through travel restrictions.
“Given how many times the introductions happened, trying to restrict traffic or movement of people obviously isn’t a solution,” Dr Andersen said. “Focusing on disease prevention and mosquito control in endemic areas is likely to be a much more successful strategy.”
When the virus did spread, the researchers found that splitting Miami into designated Zika zones—often done by neighborhood or city block—didn’t accurately represent how the virus was moving.
Within each Zika zone, the researchers discovered a mixing of multiple Zika lineages, suggesting the virus wasn’t well-confined, likely moving around with infected people.
Drs Andersen and Grubaugh hope these lessons from the 2016 epidemic will help researchers and health officials respond even faster to prevent Zika’s spread in 2017.
Behind the data
Understanding Zika’s timeline required an international team of researchers and partnerships with several health agencies.
The researchers also designed a new method of genomic sequencing just to study the Zika virus. Because Zika is hard to collect in the blood of those infected, it was a challenge for the researchers to isolate enough of its genetic material for sequencing.
To solve this problem, the researchers developed 2 different protocols to break apart the genetic material they could find and reassemble it in a useful way for analysis.
With these new protocols, the researchers sequenced the virus from 28 of the reported 256 Zika cases in Florida, as well as 7 mosquito pools, to model what happened in the larger patient group.
As they worked, the researchers released their data immediately publicly to help other researchers. They hope to release more data—and analysis—in real time as cases mount in 2017.
The research was supported by the National Institutes of Health, The Pew Charitable Trusts, The Ray Thomas Foundation, a Mahan Postdoctoral Fellowship from the Computational Biology Program at Fred Hutchinson Cancer Research Center, the US Centers for Disease Control and Prevention, the European Union Seventh Framework Programme, the United States Agency for International Development Emerging Pandemic Threats Program-2 PREDICT-2, and the Defense Advanced Research Projects Agency. ![]()
A study published in Nature appears to explain how Zika virus entered the US via Florida in 2016 and how the virus might re-enter the country this year.
By sequencing the Zika virus genome at different points in the outbreak, researchers created a family tree showing where cases originated and how quickly they spread.
The team discovered that transmission of Zika virus began in Florida at least 4—and potentially up to 40—times last year.
The researchers also traced most of the Zika lineages back to strains of the virus in the Caribbean.
“Without these genomes, we wouldn’t be able to reconstruct the history of how the virus moved around,” said study author Kristian G. Andersen, PhD, of The Scripps Research Institute in La Jolla, California.
“Rapid viral genome sequencing during ongoing outbreaks is a new development that has only been made possible over the last couple of years.”
Why Miami?
By sequencing Zika virus genomes from humans and mosquitoes—and analyzing travel and mosquito abundance data—the researchers found that several factors created a “perfect storm” for the spread of Zika virus in Miami.
“This study shows why Miami is special,” said study author Nathan D. Grubaugh, PhD, of The Scripps Research Institute.
First, Dr Grubaugh explained, Miami is home to year-round populations of Aedes aegypti mosquitoes, the main species that transmits Zika virus.
The area is also a significant travel hub, bringing in more international air and sea traffic than any other city in the continental US in 2016. Finally, Miami is an especially popular destination for travelers who have visited Zika-afflicted areas.
The researchers found that travel from the Caribbean islands may have significantly contributed to cases of Zika reaching Miami.
Of the 5.7 million international travelers entering Miami by flights and cruise ships between January and June of 2016, more than half arrived from the Caribbean.
Killing mosquitos produces results
The researchers believe Zika virus may have started transmission in Miami up to 40 times, but most travel-related cases did not lead to any secondary infections locally. The virus was more likely to reach a dead end than keep spreading.
The researchers found that one reason for the dead-ends was a direct connection between mosquito control efforts and disease prevention.
“We show that if you decrease the mosquito population in an area, the number of Zika infections goes down proportionally,” Dr Andersen said. “This means we can significantly limit the risk of Zika virus by focusing on mosquito control. This is not too surprising, but it’s important to show that there is an almost perfect correlation between the number of mosquitos and the number of human infections.”
Based on data from the outbreak, the researchers see potential in stopping the virus through mosquito control efforts in the US and other infected countries, instead of, for example, through travel restrictions.
“Given how many times the introductions happened, trying to restrict traffic or movement of people obviously isn’t a solution,” Dr Andersen said. “Focusing on disease prevention and mosquito control in endemic areas is likely to be a much more successful strategy.”
When the virus did spread, the researchers found that splitting Miami into designated Zika zones—often done by neighborhood or city block—didn’t accurately represent how the virus was moving.
Within each Zika zone, the researchers discovered a mixing of multiple Zika lineages, suggesting the virus wasn’t well-confined, likely moving around with infected people.
Drs Andersen and Grubaugh hope these lessons from the 2016 epidemic will help researchers and health officials respond even faster to prevent Zika’s spread in 2017.
Behind the data
Understanding Zika’s timeline required an international team of researchers and partnerships with several health agencies.
The researchers also designed a new method of genomic sequencing just to study the Zika virus. Because Zika is hard to collect in the blood of those infected, it was a challenge for the researchers to isolate enough of its genetic material for sequencing.
To solve this problem, the researchers developed 2 different protocols to break apart the genetic material they could find and reassemble it in a useful way for analysis.
With these new protocols, the researchers sequenced the virus from 28 of the reported 256 Zika cases in Florida, as well as 7 mosquito pools, to model what happened in the larger patient group.
As they worked, the researchers released their data immediately publicly to help other researchers. They hope to release more data—and analysis—in real time as cases mount in 2017.
The research was supported by the National Institutes of Health, The Pew Charitable Trusts, The Ray Thomas Foundation, a Mahan Postdoctoral Fellowship from the Computational Biology Program at Fred Hutchinson Cancer Research Center, the US Centers for Disease Control and Prevention, the European Union Seventh Framework Programme, the United States Agency for International Development Emerging Pandemic Threats Program-2 PREDICT-2, and the Defense Advanced Research Projects Agency. ![]()
AstraZeneca recalls lot of Brilinta in US
AstraZeneca has announced a voluntary recall of 1 lot of professional (physician) sample bottles containing 8 tablets of Brilinta® (ticagrelor).
This recall follows a report that a professional sample bottle containing 8 tablets of Brilinta 90 mg also contained Zurampic® (lesinurad) 200 mg tablets, which is also manufactured by AstraZeneca.
The company said this precautionary measure is limited to 1 lot of Brilinta (Brilinta lot #JB5047) distributed to physicians in the US between March and April of 2017.
Other forms and dosage strengths of Brilinta, including medicine obtained via US retail or mail order pharmacies, are not affected by this recall. And this recall does not affect Zurampic.
Potential risks
Unintentional dosing with Zurampic has the potential to lead to adverse renal effects, including acute renal failure, which is more common when Zurampic is given alone, as it should be used in combination with a xanthine oxidase inhibitor.
Missed doses of Brilinta increase the risk of heart attack and stroke. People with a stent who miss doses of Brilinta have a higher risk of stent thrombosis, heart attack, and death. Patients should not stop taking Brilinta without talking to their prescribing doctor.
To date, AstraZeneca has not received any reports of adverse events related to this recall.
Next steps
AstraZeneca is notifying physicians by recall letter and is arranging for the return of all recalled products. Consumers who have medicine that is being recalled should contact their physician.
Consumers with questions regarding this recall can contact the AstraZeneca Information Center at 1-800-236-9933 between the hours of 8 am and 6 pm (Eastern time) Monday to Friday, excluding holidays.
Consumers should contact their physician or healthcare provider if they have experienced any problems that may be related to taking or using Brilinta.
Adverse reactions or quality problems related to Brilinta may also be reported to the FDA’s MedWatch Adverse Event Reporting Program.
About Brilinta and Zurampic
Brilinta is intended for use to reduce the rate of cardiovascular death, heart attack, and stroke in patients with acute coronary syndrome or a history of heart attack.
Brilinta is also intended to reduce the rate of stent thrombosis in patients who have been stented for treatment of acute coronary syndrome.
Zurampic is used together with a xanthine oxidase inhibitor, such as allopurinol or Uloric, in adults with gout who still have a high uric acid level.
Brilinta 90 mg tablets are supplied as a round, biconvex, yellow, film-coated tablet, and imprinted with a “90” above a “T” on one side of the pill.
Zurampic tablets 200 mg are blue in color and elliptical/oval in shape. They are imprinted with “LES200” on one side of the pill. ![]()
AstraZeneca has announced a voluntary recall of 1 lot of professional (physician) sample bottles containing 8 tablets of Brilinta® (ticagrelor).
This recall follows a report that a professional sample bottle containing 8 tablets of Brilinta 90 mg also contained Zurampic® (lesinurad) 200 mg tablets, which is also manufactured by AstraZeneca.
The company said this precautionary measure is limited to 1 lot of Brilinta (Brilinta lot #JB5047) distributed to physicians in the US between March and April of 2017.
Other forms and dosage strengths of Brilinta, including medicine obtained via US retail or mail order pharmacies, are not affected by this recall. And this recall does not affect Zurampic.
Potential risks
Unintentional dosing with Zurampic has the potential to lead to adverse renal effects, including acute renal failure, which is more common when Zurampic is given alone, as it should be used in combination with a xanthine oxidase inhibitor.
Missed doses of Brilinta increase the risk of heart attack and stroke. People with a stent who miss doses of Brilinta have a higher risk of stent thrombosis, heart attack, and death. Patients should not stop taking Brilinta without talking to their prescribing doctor.
To date, AstraZeneca has not received any reports of adverse events related to this recall.
Next steps
AstraZeneca is notifying physicians by recall letter and is arranging for the return of all recalled products. Consumers who have medicine that is being recalled should contact their physician.
Consumers with questions regarding this recall can contact the AstraZeneca Information Center at 1-800-236-9933 between the hours of 8 am and 6 pm (Eastern time) Monday to Friday, excluding holidays.
Consumers should contact their physician or healthcare provider if they have experienced any problems that may be related to taking or using Brilinta.
Adverse reactions or quality problems related to Brilinta may also be reported to the FDA’s MedWatch Adverse Event Reporting Program.
About Brilinta and Zurampic
Brilinta is intended for use to reduce the rate of cardiovascular death, heart attack, and stroke in patients with acute coronary syndrome or a history of heart attack.
Brilinta is also intended to reduce the rate of stent thrombosis in patients who have been stented for treatment of acute coronary syndrome.
Zurampic is used together with a xanthine oxidase inhibitor, such as allopurinol or Uloric, in adults with gout who still have a high uric acid level.
Brilinta 90 mg tablets are supplied as a round, biconvex, yellow, film-coated tablet, and imprinted with a “90” above a “T” on one side of the pill.
Zurampic tablets 200 mg are blue in color and elliptical/oval in shape. They are imprinted with “LES200” on one side of the pill. ![]()
AstraZeneca has announced a voluntary recall of 1 lot of professional (physician) sample bottles containing 8 tablets of Brilinta® (ticagrelor).
This recall follows a report that a professional sample bottle containing 8 tablets of Brilinta 90 mg also contained Zurampic® (lesinurad) 200 mg tablets, which is also manufactured by AstraZeneca.
The company said this precautionary measure is limited to 1 lot of Brilinta (Brilinta lot #JB5047) distributed to physicians in the US between March and April of 2017.
Other forms and dosage strengths of Brilinta, including medicine obtained via US retail or mail order pharmacies, are not affected by this recall. And this recall does not affect Zurampic.
Potential risks
Unintentional dosing with Zurampic has the potential to lead to adverse renal effects, including acute renal failure, which is more common when Zurampic is given alone, as it should be used in combination with a xanthine oxidase inhibitor.
Missed doses of Brilinta increase the risk of heart attack and stroke. People with a stent who miss doses of Brilinta have a higher risk of stent thrombosis, heart attack, and death. Patients should not stop taking Brilinta without talking to their prescribing doctor.
To date, AstraZeneca has not received any reports of adverse events related to this recall.
Next steps
AstraZeneca is notifying physicians by recall letter and is arranging for the return of all recalled products. Consumers who have medicine that is being recalled should contact their physician.
Consumers with questions regarding this recall can contact the AstraZeneca Information Center at 1-800-236-9933 between the hours of 8 am and 6 pm (Eastern time) Monday to Friday, excluding holidays.
Consumers should contact their physician or healthcare provider if they have experienced any problems that may be related to taking or using Brilinta.
Adverse reactions or quality problems related to Brilinta may also be reported to the FDA’s MedWatch Adverse Event Reporting Program.
About Brilinta and Zurampic
Brilinta is intended for use to reduce the rate of cardiovascular death, heart attack, and stroke in patients with acute coronary syndrome or a history of heart attack.
Brilinta is also intended to reduce the rate of stent thrombosis in patients who have been stented for treatment of acute coronary syndrome.
Zurampic is used together with a xanthine oxidase inhibitor, such as allopurinol or Uloric, in adults with gout who still have a high uric acid level.
Brilinta 90 mg tablets are supplied as a round, biconvex, yellow, film-coated tablet, and imprinted with a “90” above a “T” on one side of the pill.
Zurampic tablets 200 mg are blue in color and elliptical/oval in shape. They are imprinted with “LES200” on one side of the pill. ![]()
Finding could lead to ‘Holy Grail’ of antithrombotic therapy
The protein CIB1 may be a superior target for antithrombotic therapy, according to researchers.
The team found that CIB1 plays a role in thrombus development but not in initial clot formation.
“The Holy Grail of our field is to reduce unwanted thrombus formation without completely blocking other important platelet functions,” said study author Ulhas Naik, PhD, of Thomas Jefferson University in Philadelphia, Pennsylvania.
He and his colleagues believe targeting CIB1 may do just that.
In earlier work, the researchers showed that CIB1 was involved in thrombus formation. They found mice that lacked the CIB1 gene were less likely to form a thrombus.
That work also indicated that mice lacking the CIB1 were still able to form a platelet plug, suggesting this gene was involved only in the process of thrombus formation.
To investigate this possibility further and to demonstrate that the process was relevant to human physiology, Dr Naik and his colleagues conducted the current study.
The team described this work in PLOS ONE.
The researchers studied human platelets and probed the molecules that interacted with CIB1 at different time points after platelet activation.
The team found that CIB1 does not begin to bind and interact with platelet molecular machinery until after filopodia formation, which allows platelets to cross-link to one another and begin to form a plug.
The study also elucidated a number of molecules that CIB1 interacts with during outside-in signaling and thrombus formation.
“This work demonstrates that CIB1 could be a good antithrombotic drug target,” Dr Naik said. “If we block CIB1, it hampers thrombus formation without interfering with platelet plug formation. If developed further, blocking CIB1 could reduce the risk of heart attack and stroke without increasing the risk for excessive bleeding that is the trade-off of current medication.”
The next step for Dr Naik and his colleagues is screening for small-molecule compounds that would inhibit CIB1 and could be developed into new therapies. ![]()
The protein CIB1 may be a superior target for antithrombotic therapy, according to researchers.
The team found that CIB1 plays a role in thrombus development but not in initial clot formation.
“The Holy Grail of our field is to reduce unwanted thrombus formation without completely blocking other important platelet functions,” said study author Ulhas Naik, PhD, of Thomas Jefferson University in Philadelphia, Pennsylvania.
He and his colleagues believe targeting CIB1 may do just that.
In earlier work, the researchers showed that CIB1 was involved in thrombus formation. They found mice that lacked the CIB1 gene were less likely to form a thrombus.
That work also indicated that mice lacking the CIB1 were still able to form a platelet plug, suggesting this gene was involved only in the process of thrombus formation.
To investigate this possibility further and to demonstrate that the process was relevant to human physiology, Dr Naik and his colleagues conducted the current study.
The team described this work in PLOS ONE.
The researchers studied human platelets and probed the molecules that interacted with CIB1 at different time points after platelet activation.
The team found that CIB1 does not begin to bind and interact with platelet molecular machinery until after filopodia formation, which allows platelets to cross-link to one another and begin to form a plug.
The study also elucidated a number of molecules that CIB1 interacts with during outside-in signaling and thrombus formation.
“This work demonstrates that CIB1 could be a good antithrombotic drug target,” Dr Naik said. “If we block CIB1, it hampers thrombus formation without interfering with platelet plug formation. If developed further, blocking CIB1 could reduce the risk of heart attack and stroke without increasing the risk for excessive bleeding that is the trade-off of current medication.”
The next step for Dr Naik and his colleagues is screening for small-molecule compounds that would inhibit CIB1 and could be developed into new therapies. ![]()
The protein CIB1 may be a superior target for antithrombotic therapy, according to researchers.
The team found that CIB1 plays a role in thrombus development but not in initial clot formation.
“The Holy Grail of our field is to reduce unwanted thrombus formation without completely blocking other important platelet functions,” said study author Ulhas Naik, PhD, of Thomas Jefferson University in Philadelphia, Pennsylvania.
He and his colleagues believe targeting CIB1 may do just that.
In earlier work, the researchers showed that CIB1 was involved in thrombus formation. They found mice that lacked the CIB1 gene were less likely to form a thrombus.
That work also indicated that mice lacking the CIB1 were still able to form a platelet plug, suggesting this gene was involved only in the process of thrombus formation.
To investigate this possibility further and to demonstrate that the process was relevant to human physiology, Dr Naik and his colleagues conducted the current study.
The team described this work in PLOS ONE.
The researchers studied human platelets and probed the molecules that interacted with CIB1 at different time points after platelet activation.
The team found that CIB1 does not begin to bind and interact with platelet molecular machinery until after filopodia formation, which allows platelets to cross-link to one another and begin to form a plug.
The study also elucidated a number of molecules that CIB1 interacts with during outside-in signaling and thrombus formation.
“This work demonstrates that CIB1 could be a good antithrombotic drug target,” Dr Naik said. “If we block CIB1, it hampers thrombus formation without interfering with platelet plug formation. If developed further, blocking CIB1 could reduce the risk of heart attack and stroke without increasing the risk for excessive bleeding that is the trade-off of current medication.”
The next step for Dr Naik and his colleagues is screening for small-molecule compounds that would inhibit CIB1 and could be developed into new therapies.
Study supports wider use of dried blood samples
Researchers have found evidence to suggest that dried blood samples may sometimes be a suitable alternative to conventional blood sampling.
The team measured levels of 92 proteins in millimeter-sized circles punched out of dried blood samples.
They found that, in many cases, little happens to these proteins when they are allowed to dry.
Most of the proteins remain unaltered after 30 years, or they change only minimally.
However, the proteins can be affected by storage temperatures.
Still, the researchers believe these results suggest dried blood samples could be used more widely—for routine health checks and to set up large-scale biobanks, with patients collecting the blood samples themselves.
“[Y]ou can prick your own finger and send in a dried blood spot by post,” study author Ulf Landegren, MD, PhD, of Uppsala University in Sweden.
“[A]t a minimal cost, it will be possible to build gigantic biobanks of samples obtained on a routine clinical basis. This means that samples can be taken before the clinical debut of a disease to identify markers of value for early diagnosis, improving the scope for curative treatment.”
Dr Landegren and his colleagues discussed these possibilities in a paper published in Molecular and Cellular Proteomics.
The researchers analyzed dried blood samples, measuring levels of 92 proteins that are relevant in oncology. To determine the effects of long-term storage, the team examined what happens to protein detection as an effect of the drying process.
Some of the dried blood samples analyzed had been collected recently, while others had been preserved for up to 30 years in biobanks in Sweden and Denmark. These 2 biobanks keep their dried blood samples at different temperatures: the Swedish one at +4°C and the Danish one at -24°C.
The researchers also looked at wet plasma samples kept at -70°C for corresponding periods of time.
“Our conclusion is that we can measure levels of 92 proteins with very high precision and sensitivity using PEA [proximity extension assay] technology in the tiny, punched-out discs from a dried blood spot,” said study author Johan Björkesten, a doctoral student at Uppsala University.
“The actual drying process has a negligible effect on the various proteins, and the effect is reproducible, which means that it can be included in the calculation.”
The researchers did find that long-term storage affects the detectability of certain proteins more than others.
Most proteins remain completely intact after 30 years or exhibit minimal changes. However, levels of some proteins decrease so that half the quantity remains after a period of between 10 and 50 years.
The researchers also found that a relatively low storage temperature is preferable for proteins that are affected by storage.
Protein detection was less affected when dried blood samples were stored at -24°C than when they were stored at +4°C. Over the 30-year period, detectability was not affected for 34% of proteins stored at +4°C and 76% of proteins stored at -24°C.
However, storing wet plasma at -70°C preserved proteins better than dried blood sample storage at -24°C. Detectability decreased for 5% of the proteins stored wet at -70°C for 45 years, compared to 24% for proteins in dried samples stored at -24°C for 30 years.
The researchers did note, though, that this part of their analysis was complicated by some confounding factors, so this was not a clear, direct comparison between wet and dry samples.
Researchers have found evidence to suggest that dried blood samples may sometimes be a suitable alternative to conventional blood sampling.
The team measured levels of 92 proteins in millimeter-sized circles punched out of dried blood samples.
They found that, in many cases, little happens to these proteins when they are allowed to dry.
Most of the proteins remain unaltered after 30 years, or they change only minimally.
However, the proteins can be affected by storage temperatures.
Still, the researchers believe these results suggest dried blood samples could be used more widely—for routine health checks and to set up large-scale biobanks, with patients collecting the blood samples themselves.
“[Y]ou can prick your own finger and send in a dried blood spot by post,” study author Ulf Landegren, MD, PhD, of Uppsala University in Sweden.
“[A]t a minimal cost, it will be possible to build gigantic biobanks of samples obtained on a routine clinical basis. This means that samples can be taken before the clinical debut of a disease to identify markers of value for early diagnosis, improving the scope for curative treatment.”
Dr Landegren and his colleagues discussed these possibilities in a paper published in Molecular and Cellular Proteomics.
The researchers analyzed dried blood samples, measuring levels of 92 proteins that are relevant in oncology. To determine the effects of long-term storage, the team examined what happens to protein detection as an effect of the drying process.
Some of the dried blood samples analyzed had been collected recently, while others had been preserved for up to 30 years in biobanks in Sweden and Denmark. These 2 biobanks keep their dried blood samples at different temperatures: the Swedish one at +4°C and the Danish one at -24°C.
The researchers also looked at wet plasma samples kept at -70°C for corresponding periods of time.
“Our conclusion is that we can measure levels of 92 proteins with very high precision and sensitivity using PEA [proximity extension assay] technology in the tiny, punched-out discs from a dried blood spot,” said study author Johan Björkesten, a doctoral student at Uppsala University.
“The actual drying process has a negligible effect on the various proteins, and the effect is reproducible, which means that it can be included in the calculation.”
The researchers did find that long-term storage affects the detectability of certain proteins more than others.
Most proteins remain completely intact after 30 years or exhibit minimal changes. However, levels of some proteins decrease so that half the quantity remains after a period of between 10 and 50 years.
The researchers also found that a relatively low storage temperature is preferable for proteins that are affected by storage.
Protein detection was less affected when dried blood samples were stored at -24°C than when they were stored at +4°C. Over the 30-year period, detectability was not affected for 34% of proteins stored at +4°C and 76% of proteins stored at -24°C.
However, storing wet plasma at -70°C preserved proteins better than dried blood sample storage at -24°C. Detectability decreased for 5% of the proteins stored wet at -70°C for 45 years, compared to 24% for proteins in dried samples stored at -24°C for 30 years.
The researchers did note, though, that this part of their analysis was complicated by some confounding factors, so this was not a clear, direct comparison between wet and dry samples.
Researchers have found evidence to suggest that dried blood samples may sometimes be a suitable alternative to conventional blood sampling.
The team measured levels of 92 proteins in millimeter-sized circles punched out of dried blood samples.
They found that, in many cases, little happens to these proteins when they are allowed to dry.
Most of the proteins remain unaltered after 30 years, or they change only minimally.
However, the proteins can be affected by storage temperatures.
Still, the researchers believe these results suggest dried blood samples could be used more widely—for routine health checks and to set up large-scale biobanks, with patients collecting the blood samples themselves.
“[Y]ou can prick your own finger and send in a dried blood spot by post,” study author Ulf Landegren, MD, PhD, of Uppsala University in Sweden.
“[A]t a minimal cost, it will be possible to build gigantic biobanks of samples obtained on a routine clinical basis. This means that samples can be taken before the clinical debut of a disease to identify markers of value for early diagnosis, improving the scope for curative treatment.”
Dr Landegren and his colleagues discussed these possibilities in a paper published in Molecular and Cellular Proteomics.
The researchers analyzed dried blood samples, measuring levels of 92 proteins that are relevant in oncology. To determine the effects of long-term storage, the team examined what happens to protein detection as an effect of the drying process.
Some of the dried blood samples analyzed had been collected recently, while others had been preserved for up to 30 years in biobanks in Sweden and Denmark. These 2 biobanks keep their dried blood samples at different temperatures: the Swedish one at +4°C and the Danish one at -24°C.
The researchers also looked at wet plasma samples kept at -70°C for corresponding periods of time.
“Our conclusion is that we can measure levels of 92 proteins with very high precision and sensitivity using PEA [proximity extension assay] technology in the tiny, punched-out discs from a dried blood spot,” said study author Johan Björkesten, a doctoral student at Uppsala University.
“The actual drying process has a negligible effect on the various proteins, and the effect is reproducible, which means that it can be included in the calculation.”
The researchers did find that long-term storage affects the detectability of certain proteins more than others.
Most proteins remain completely intact after 30 years or exhibit minimal changes. However, levels of some proteins decrease so that half the quantity remains after a period of between 10 and 50 years.
The researchers also found that a relatively low storage temperature is preferable for proteins that are affected by storage.
Protein detection was less affected when dried blood samples were stored at -24°C than when they were stored at +4°C. Over the 30-year period, detectability was not affected for 34% of proteins stored at +4°C and 76% of proteins stored at -24°C.
However, storing wet plasma at -70°C preserved proteins better than dried blood sample storage at -24°C. Detectability decreased for 5% of the proteins stored wet at -70°C for 45 years, compared to 24% for proteins in dried samples stored at -24°C for 30 years.
The researchers did note, though, that this part of their analysis was complicated by some confounding factors, so this was not a clear, direct comparison between wet and dry samples.
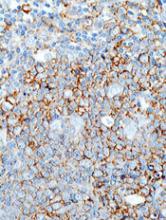
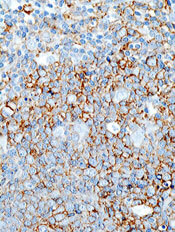
EC grants drug orphan designation for AML
The European Commission (EC) has granted orphan designation to GMI-1271 for the treatment of acute myeloid leukemia (AML).
GMI-1271 is an E-selectin antagonist being developed by GlycoMimetics, Inc.
The product also has orphan designation, fast track designation, and breakthrough therapy designation in the US.
GMI-1271 is currently being evaluated in a phase 1/2 trial of patients with relapsed or refractory AML and patients age 60 and older with newly diagnosed AML.
The patients are receiving GM-1271 in combination with chemotherapy. The relapsed/refractory group is receiving mitoxantrone, etoposide, and cytarabine. The newly diagnosed patients are receiving cytarabine and idarubicin (7+3).
GlycoMimetics plans to present data from this trial at the 2017 American Society of Clinical Oncology (ASCO) Annual Meeting as abstracts 2520 and 2560.
The company also plans to present the research at the 22nd Congress of the European Hematology Association (EHA) as abstracts P547 and P203.
About orphan designation
Orphan designation provides regulatory and financial incentives for companies to develop and market therapies that treat life-threatening or chronically debilitating conditions affecting no more than 5 in 10,000 people in the European Union, and where no satisfactory treatment is available.
Orphan designation provides a 10-year period of marketing exclusivity if the drug receives regulatory approval.
The designation also provides incentives for companies seeking protocol assistance from the European Medicines Agency during the product development phase and direct access to the centralized authorization procedure.
The European Medicines Agency adopts an opinion on the granting of orphan drug designation, and that opinion is submitted to the EC for a final decision. The EC typically makes a decision within 30 days of that submission.
The European Commission (EC) has granted orphan designation to GMI-1271 for the treatment of acute myeloid leukemia (AML).
GMI-1271 is an E-selectin antagonist being developed by GlycoMimetics, Inc.
The product also has orphan designation, fast track designation, and breakthrough therapy designation in the US.
GMI-1271 is currently being evaluated in a phase 1/2 trial of patients with relapsed or refractory AML and patients age 60 and older with newly diagnosed AML.
The patients are receiving GM-1271 in combination with chemotherapy. The relapsed/refractory group is receiving mitoxantrone, etoposide, and cytarabine. The newly diagnosed patients are receiving cytarabine and idarubicin (7+3).
GlycoMimetics plans to present data from this trial at the 2017 American Society of Clinical Oncology (ASCO) Annual Meeting as abstracts 2520 and 2560.
The company also plans to present the research at the 22nd Congress of the European Hematology Association (EHA) as abstracts P547 and P203.
About orphan designation
Orphan designation provides regulatory and financial incentives for companies to develop and market therapies that treat life-threatening or chronically debilitating conditions affecting no more than 5 in 10,000 people in the European Union, and where no satisfactory treatment is available.
Orphan designation provides a 10-year period of marketing exclusivity if the drug receives regulatory approval.
The designation also provides incentives for companies seeking protocol assistance from the European Medicines Agency during the product development phase and direct access to the centralized authorization procedure.
The European Medicines Agency adopts an opinion on the granting of orphan drug designation, and that opinion is submitted to the EC for a final decision. The EC typically makes a decision within 30 days of that submission.
The European Commission (EC) has granted orphan designation to GMI-1271 for the treatment of acute myeloid leukemia (AML).
GMI-1271 is an E-selectin antagonist being developed by GlycoMimetics, Inc.
The product also has orphan designation, fast track designation, and breakthrough therapy designation in the US.
GMI-1271 is currently being evaluated in a phase 1/2 trial of patients with relapsed or refractory AML and patients age 60 and older with newly diagnosed AML.
The patients are receiving GM-1271 in combination with chemotherapy. The relapsed/refractory group is receiving mitoxantrone, etoposide, and cytarabine. The newly diagnosed patients are receiving cytarabine and idarubicin (7+3).
GlycoMimetics plans to present data from this trial at the 2017 American Society of Clinical Oncology (ASCO) Annual Meeting as abstracts 2520 and 2560.
The company also plans to present the research at the 22nd Congress of the European Hematology Association (EHA) as abstracts P547 and P203.
About orphan designation
Orphan designation provides regulatory and financial incentives for companies to develop and market therapies that treat life-threatening or chronically debilitating conditions affecting no more than 5 in 10,000 people in the European Union, and where no satisfactory treatment is available.
Orphan designation provides a 10-year period of marketing exclusivity if the drug receives regulatory approval.
The designation also provides incentives for companies seeking protocol assistance from the European Medicines Agency during the product development phase and direct access to the centralized authorization procedure.
The European Medicines Agency adopts an opinion on the granting of orphan drug designation, and that opinion is submitted to the EC for a final decision. The EC typically makes a decision within 30 days of that submission.
Group creates ‘authentic’ HSCs from endothelial cells
Researchers say they have found a way to convert adult mouse endothelial cells into “authentic” hematopoietic stem cells (HSCs).
The team says these HSCs have a transcriptome and long-term self-renewal capacity that are similar to those of adult HSCs that are produced naturally.
In addition, the lab-generated HSCs were capable of engraftment and multi-lineage reconstitution in mice.
“This is a game-changing breakthrough that brings us closer not only to treat blood disorders, but also to deciphering the complex biology of stem-cell self-renewal machinery,” said Shahin Rafii, MD, of Weill Cornell Medicine in New York, New York.
“This is exciting because it provides us with a path towards generating clinically useful quantities of normal stem cells for transplantation that may help us cure patients with genetic and acquired blood diseases,” added Joseph Scandura, MD, PhD, also of Weill Cornell Medicine.
Drs Scandura and Rafii and their colleagues described this research in Nature.
The researchers took vascular endothelial cells from adult mice and induced expression of the transcription-factor-encoding genes Fosb, Gfi1, Runx1, and Spi1.
The cells were grown and multiplied in co-culture with an engineered vascular niche.
This produced HSCs that were transplanted into irradiated mice.
The researchers said these HSCs were capable of long-term engraftment and hematopoietic reconstitution of myelopoiesis and both innate and adaptive immune function.
In addition, the HSCs were endowed with the same genetic attributes as normal adult HSCs.
If this method of generating HSCs in the lab can be scaled up and applied to humans, it could have wide-ranging clinical implications, according to the researchers.
“It might allow us to provide healthy stem cells to patients who need bone marrow donors but have no genetic match,” Dr Scandura said. “It could lead to new ways to cure leukemia and may help us correct genetic defects that cause blood diseases like sickle cell anemia.”
“More importantly, our vascular niche stem cell expansion model may be employed to clone the key unknown growth factors produced by this niche that are essential for self-perpetuation of stem cells,” Dr Rafii said. “Identification of those factors could be important for unraveling the secrets of stem cells’ longevity and translating the potential of stem cell therapy to the clinical setting.”
Researchers say they have found a way to convert adult mouse endothelial cells into “authentic” hematopoietic stem cells (HSCs).
The team says these HSCs have a transcriptome and long-term self-renewal capacity that are similar to those of adult HSCs that are produced naturally.
In addition, the lab-generated HSCs were capable of engraftment and multi-lineage reconstitution in mice.
“This is a game-changing breakthrough that brings us closer not only to treat blood disorders, but also to deciphering the complex biology of stem-cell self-renewal machinery,” said Shahin Rafii, MD, of Weill Cornell Medicine in New York, New York.
“This is exciting because it provides us with a path towards generating clinically useful quantities of normal stem cells for transplantation that may help us cure patients with genetic and acquired blood diseases,” added Joseph Scandura, MD, PhD, also of Weill Cornell Medicine.
Drs Scandura and Rafii and their colleagues described this research in Nature.
The researchers took vascular endothelial cells from adult mice and induced expression of the transcription-factor-encoding genes Fosb, Gfi1, Runx1, and Spi1.
The cells were grown and multiplied in co-culture with an engineered vascular niche.
This produced HSCs that were transplanted into irradiated mice.
The researchers said these HSCs were capable of long-term engraftment and hematopoietic reconstitution of myelopoiesis and both innate and adaptive immune function.
In addition, the HSCs were endowed with the same genetic attributes as normal adult HSCs.
If this method of generating HSCs in the lab can be scaled up and applied to humans, it could have wide-ranging clinical implications, according to the researchers.
“It might allow us to provide healthy stem cells to patients who need bone marrow donors but have no genetic match,” Dr Scandura said. “It could lead to new ways to cure leukemia and may help us correct genetic defects that cause blood diseases like sickle cell anemia.”
“More importantly, our vascular niche stem cell expansion model may be employed to clone the key unknown growth factors produced by this niche that are essential for self-perpetuation of stem cells,” Dr Rafii said. “Identification of those factors could be important for unraveling the secrets of stem cells’ longevity and translating the potential of stem cell therapy to the clinical setting.”
Researchers say they have found a way to convert adult mouse endothelial cells into “authentic” hematopoietic stem cells (HSCs).
The team says these HSCs have a transcriptome and long-term self-renewal capacity that are similar to those of adult HSCs that are produced naturally.
In addition, the lab-generated HSCs were capable of engraftment and multi-lineage reconstitution in mice.
“This is a game-changing breakthrough that brings us closer not only to treat blood disorders, but also to deciphering the complex biology of stem-cell self-renewal machinery,” said Shahin Rafii, MD, of Weill Cornell Medicine in New York, New York.
“This is exciting because it provides us with a path towards generating clinically useful quantities of normal stem cells for transplantation that may help us cure patients with genetic and acquired blood diseases,” added Joseph Scandura, MD, PhD, also of Weill Cornell Medicine.
Drs Scandura and Rafii and their colleagues described this research in Nature.
The researchers took vascular endothelial cells from adult mice and induced expression of the transcription-factor-encoding genes Fosb, Gfi1, Runx1, and Spi1.
The cells were grown and multiplied in co-culture with an engineered vascular niche.
This produced HSCs that were transplanted into irradiated mice.
The researchers said these HSCs were capable of long-term engraftment and hematopoietic reconstitution of myelopoiesis and both innate and adaptive immune function.
In addition, the HSCs were endowed with the same genetic attributes as normal adult HSCs.
If this method of generating HSCs in the lab can be scaled up and applied to humans, it could have wide-ranging clinical implications, according to the researchers.
“It might allow us to provide healthy stem cells to patients who need bone marrow donors but have no genetic match,” Dr Scandura said. “It could lead to new ways to cure leukemia and may help us correct genetic defects that cause blood diseases like sickle cell anemia.”
“More importantly, our vascular niche stem cell expansion model may be employed to clone the key unknown growth factors produced by this niche that are essential for self-perpetuation of stem cells,” Dr Rafii said. “Identification of those factors could be important for unraveling the secrets of stem cells’ longevity and translating the potential of stem cell therapy to the clinical setting.”