User login
Genes may be targets for AML therapy
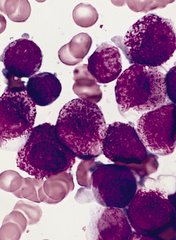
Two genes are critical to the development of acute myeloid leukemia (AML), according to research published in Cancer Cell.
Previous research suggested the genes, KDM4C and PRMT1, are key players in transcription regulation during both normal and disease development.
The new study showed that, during AML development, KDM4C and PRMT1 are recruited to enable the transformation of blood cells into cancer cells.
The genes work in tandem, and, if either is not fully active, AML does not develop.
The researchers made these discoveries by inhibiting KDM4C and PRMT1—either genetically or pharmacologically—in mice with AML.
When either gene was silenced via genetic means, the majority of the mice were still alive at the end of the researchers’ 60-day experiment. However, the majority of control mice died in less than 40 days.
The team observed similarly favorable results when they inhibited either gene with drugs—the PRMT1 inhibitor AMI-408 and the KDM4C inhibitor SD70.
The median disease latency was 48 days in mice that received AMI-408 and 36 days in control mice. The median disease latency was 62 days in mice that received SD70 and 55 days in control mice.
“The demonstration of how critical these genes are to cancer transformation and treatment could be highly significant for the design of new drugs,” said study author Eric So, PhD, of King’s College London in the UK.
“Further work is needed to develop and refine drugs to maximize their effects and so that they are suitable for patients. Clinical trials will then be needed to see how leukemia patients respond to these drugs and how use of them can be optimized.” ![]()

Two genes are critical to the development of acute myeloid leukemia (AML), according to research published in Cancer Cell.
Previous research suggested the genes, KDM4C and PRMT1, are key players in transcription regulation during both normal and disease development.
The new study showed that, during AML development, KDM4C and PRMT1 are recruited to enable the transformation of blood cells into cancer cells.
The genes work in tandem, and, if either is not fully active, AML does not develop.
The researchers made these discoveries by inhibiting KDM4C and PRMT1—either genetically or pharmacologically—in mice with AML.
When either gene was silenced via genetic means, the majority of the mice were still alive at the end of the researchers’ 60-day experiment. However, the majority of control mice died in less than 40 days.
The team observed similarly favorable results when they inhibited either gene with drugs—the PRMT1 inhibitor AMI-408 and the KDM4C inhibitor SD70.
The median disease latency was 48 days in mice that received AMI-408 and 36 days in control mice. The median disease latency was 62 days in mice that received SD70 and 55 days in control mice.
“The demonstration of how critical these genes are to cancer transformation and treatment could be highly significant for the design of new drugs,” said study author Eric So, PhD, of King’s College London in the UK.
“Further work is needed to develop and refine drugs to maximize their effects and so that they are suitable for patients. Clinical trials will then be needed to see how leukemia patients respond to these drugs and how use of them can be optimized.” ![]()

Two genes are critical to the development of acute myeloid leukemia (AML), according to research published in Cancer Cell.
Previous research suggested the genes, KDM4C and PRMT1, are key players in transcription regulation during both normal and disease development.
The new study showed that, during AML development, KDM4C and PRMT1 are recruited to enable the transformation of blood cells into cancer cells.
The genes work in tandem, and, if either is not fully active, AML does not develop.
The researchers made these discoveries by inhibiting KDM4C and PRMT1—either genetically or pharmacologically—in mice with AML.
When either gene was silenced via genetic means, the majority of the mice were still alive at the end of the researchers’ 60-day experiment. However, the majority of control mice died in less than 40 days.
The team observed similarly favorable results when they inhibited either gene with drugs—the PRMT1 inhibitor AMI-408 and the KDM4C inhibitor SD70.
The median disease latency was 48 days in mice that received AMI-408 and 36 days in control mice. The median disease latency was 62 days in mice that received SD70 and 55 days in control mice.
“The demonstration of how critical these genes are to cancer transformation and treatment could be highly significant for the design of new drugs,” said study author Eric So, PhD, of King’s College London in the UK.
“Further work is needed to develop and refine drugs to maximize their effects and so that they are suitable for patients. Clinical trials will then be needed to see how leukemia patients respond to these drugs and how use of them can be optimized.” ![]()
Forces driving leukemia differ in kids and adults

New research suggests childhood leukemias are forged by different evolutionary forces than leukemias in older adults.
Researchers used a computational model to characterize the population dynamics of hematopoietic stem cells (HSCs) that give rise to leukemias.
And they found the evolutionary force known as “drift” contributes to leukemia development in young children but not in older adults.
“Basically, leukemia risk early in life may be more dictated by chance than by the typical ‘survival of the fittest’ that characterizes leukemia formation in older adults,” explained study author James DeGregori, PhD, of the University of Colorado School of Medicine in Aurora.
He and his colleagues recounted this discovery in PNAS.
With previous work, the DeGregori lab showed that the inevitable tissue decline associated with aging benefits HSCs with mutations that allow the cells to better adapt to the new ecosystem.
In contrast, the ecosystem of young tissue favors healthy cells. Optimized by millions of years of co-evolution, most mutations make cells less fit for the ecosystem of young, healthy tissue and lead to purging of mutant cells from the tissue.
With the current study, Dr DeGregori and his colleagues made a surprising discovery. Despite the ability of young tissue to select against cells with cancer-causing mutations, the computational model showed increased proportions of specific, mutation-bearing HSCs in the first few years after birth.
And these mutated cells were not dependent on the effect of the mutation on cell fitness. In other words, the mutation-bearing cells were not more fit than cells without the mutations. Instead of the survival-of-the-fittest form of natural selection that drives the evolution of cancer in older adults, there was another force at work.
In fact, the researchers discovered 2 factors that influence the development of childhood leukemia: the small HSC pool size at birth and the high rate of cell division necessary for body growth early in life.
The high rate of cell division increases the risk of leukemia because mutations largely happen during cell divisions. More cell divisions mean more mutations, and this increases the risk that some of these mutations could contribute to leukemia development.
The small HSC pool size influences leukemia development via the evolutionary force known as drift. Drift is the role of chance—the possibility that, despite being less fit, an animal, organism, or HSC with an oncogenic mutation will survive to shift the genetic makeup of the population.
The influence of drift is greater in small populations and, in this case, small stem cell pools. In the small HSC pools of young children, drift becomes important as a lucky genotype may end up with a larger share of the total HSC pool than warranted by its fitness status.
If this lucky cell clone happens to have a mutation that can start the HSC down the path toward leukemia, this drift-driven expansion should increase the risk of leukemia by increasing the number of HSCs with this mutation.
“Thus, early somatic evolution in HSC pools is significantly impacted by drift, with selection playing a lesser role,” Dr DeGregori and his colleagues wrote.
On the other hand, the impact of drift lessens as the HSC pool grows along with an infant’s body to reach adult size. The larger HSC pool size decreases the role of drift in the success of particular cells in the tissue.
In addition, as the pool size reaches its maximum, the HSC division rate slows to a crawl (as these stem cells enter the maintenance rather than growth phase). With a landscape of healthy, youthful tissues and low rates of mutation due to low cell division rates, the odds of leukemia diminish.
“With a large population of healthy cells optimized to young, healthy tissue, the ability of mutations, including cancerous mutations, to drive uncontrolled cell proliferation is reduced,” Dr DeGregori said.
However, in old age, tissue decline promotes selection for adaptive mutations, leading to the expansion of potentially oncogenic HSC clones that will again increase the risk of leukemia.
Thus, this research shows that, in early life, leukemias are driven by mutation and drift. And in later life, leukemias are driven by mutation and selection.
“We show that leukemias of children and older adults are different diseases, forged by different evolutionary forces and propagated under different circumstances,” Dr DeGregori said.
He and his colleagues believe this understanding raises the possibility of a new approach to cancer treatment. Perhaps researchers could find a way to manipulate the parameters of cell evolution or manipulate the tissue ecosystem to decrease cancer risk. ![]()

New research suggests childhood leukemias are forged by different evolutionary forces than leukemias in older adults.
Researchers used a computational model to characterize the population dynamics of hematopoietic stem cells (HSCs) that give rise to leukemias.
And they found the evolutionary force known as “drift” contributes to leukemia development in young children but not in older adults.
“Basically, leukemia risk early in life may be more dictated by chance than by the typical ‘survival of the fittest’ that characterizes leukemia formation in older adults,” explained study author James DeGregori, PhD, of the University of Colorado School of Medicine in Aurora.
He and his colleagues recounted this discovery in PNAS.
With previous work, the DeGregori lab showed that the inevitable tissue decline associated with aging benefits HSCs with mutations that allow the cells to better adapt to the new ecosystem.
In contrast, the ecosystem of young tissue favors healthy cells. Optimized by millions of years of co-evolution, most mutations make cells less fit for the ecosystem of young, healthy tissue and lead to purging of mutant cells from the tissue.
With the current study, Dr DeGregori and his colleagues made a surprising discovery. Despite the ability of young tissue to select against cells with cancer-causing mutations, the computational model showed increased proportions of specific, mutation-bearing HSCs in the first few years after birth.
And these mutated cells were not dependent on the effect of the mutation on cell fitness. In other words, the mutation-bearing cells were not more fit than cells without the mutations. Instead of the survival-of-the-fittest form of natural selection that drives the evolution of cancer in older adults, there was another force at work.
In fact, the researchers discovered 2 factors that influence the development of childhood leukemia: the small HSC pool size at birth and the high rate of cell division necessary for body growth early in life.
The high rate of cell division increases the risk of leukemia because mutations largely happen during cell divisions. More cell divisions mean more mutations, and this increases the risk that some of these mutations could contribute to leukemia development.
The small HSC pool size influences leukemia development via the evolutionary force known as drift. Drift is the role of chance—the possibility that, despite being less fit, an animal, organism, or HSC with an oncogenic mutation will survive to shift the genetic makeup of the population.
The influence of drift is greater in small populations and, in this case, small stem cell pools. In the small HSC pools of young children, drift becomes important as a lucky genotype may end up with a larger share of the total HSC pool than warranted by its fitness status.
If this lucky cell clone happens to have a mutation that can start the HSC down the path toward leukemia, this drift-driven expansion should increase the risk of leukemia by increasing the number of HSCs with this mutation.
“Thus, early somatic evolution in HSC pools is significantly impacted by drift, with selection playing a lesser role,” Dr DeGregori and his colleagues wrote.
On the other hand, the impact of drift lessens as the HSC pool grows along with an infant’s body to reach adult size. The larger HSC pool size decreases the role of drift in the success of particular cells in the tissue.
In addition, as the pool size reaches its maximum, the HSC division rate slows to a crawl (as these stem cells enter the maintenance rather than growth phase). With a landscape of healthy, youthful tissues and low rates of mutation due to low cell division rates, the odds of leukemia diminish.
“With a large population of healthy cells optimized to young, healthy tissue, the ability of mutations, including cancerous mutations, to drive uncontrolled cell proliferation is reduced,” Dr DeGregori said.
However, in old age, tissue decline promotes selection for adaptive mutations, leading to the expansion of potentially oncogenic HSC clones that will again increase the risk of leukemia.
Thus, this research shows that, in early life, leukemias are driven by mutation and drift. And in later life, leukemias are driven by mutation and selection.
“We show that leukemias of children and older adults are different diseases, forged by different evolutionary forces and propagated under different circumstances,” Dr DeGregori said.
He and his colleagues believe this understanding raises the possibility of a new approach to cancer treatment. Perhaps researchers could find a way to manipulate the parameters of cell evolution or manipulate the tissue ecosystem to decrease cancer risk. ![]()

New research suggests childhood leukemias are forged by different evolutionary forces than leukemias in older adults.
Researchers used a computational model to characterize the population dynamics of hematopoietic stem cells (HSCs) that give rise to leukemias.
And they found the evolutionary force known as “drift” contributes to leukemia development in young children but not in older adults.
“Basically, leukemia risk early in life may be more dictated by chance than by the typical ‘survival of the fittest’ that characterizes leukemia formation in older adults,” explained study author James DeGregori, PhD, of the University of Colorado School of Medicine in Aurora.
He and his colleagues recounted this discovery in PNAS.
With previous work, the DeGregori lab showed that the inevitable tissue decline associated with aging benefits HSCs with mutations that allow the cells to better adapt to the new ecosystem.
In contrast, the ecosystem of young tissue favors healthy cells. Optimized by millions of years of co-evolution, most mutations make cells less fit for the ecosystem of young, healthy tissue and lead to purging of mutant cells from the tissue.
With the current study, Dr DeGregori and his colleagues made a surprising discovery. Despite the ability of young tissue to select against cells with cancer-causing mutations, the computational model showed increased proportions of specific, mutation-bearing HSCs in the first few years after birth.
And these mutated cells were not dependent on the effect of the mutation on cell fitness. In other words, the mutation-bearing cells were not more fit than cells without the mutations. Instead of the survival-of-the-fittest form of natural selection that drives the evolution of cancer in older adults, there was another force at work.
In fact, the researchers discovered 2 factors that influence the development of childhood leukemia: the small HSC pool size at birth and the high rate of cell division necessary for body growth early in life.
The high rate of cell division increases the risk of leukemia because mutations largely happen during cell divisions. More cell divisions mean more mutations, and this increases the risk that some of these mutations could contribute to leukemia development.
The small HSC pool size influences leukemia development via the evolutionary force known as drift. Drift is the role of chance—the possibility that, despite being less fit, an animal, organism, or HSC with an oncogenic mutation will survive to shift the genetic makeup of the population.
The influence of drift is greater in small populations and, in this case, small stem cell pools. In the small HSC pools of young children, drift becomes important as a lucky genotype may end up with a larger share of the total HSC pool than warranted by its fitness status.
If this lucky cell clone happens to have a mutation that can start the HSC down the path toward leukemia, this drift-driven expansion should increase the risk of leukemia by increasing the number of HSCs with this mutation.
“Thus, early somatic evolution in HSC pools is significantly impacted by drift, with selection playing a lesser role,” Dr DeGregori and his colleagues wrote.
On the other hand, the impact of drift lessens as the HSC pool grows along with an infant’s body to reach adult size. The larger HSC pool size decreases the role of drift in the success of particular cells in the tissue.
In addition, as the pool size reaches its maximum, the HSC division rate slows to a crawl (as these stem cells enter the maintenance rather than growth phase). With a landscape of healthy, youthful tissues and low rates of mutation due to low cell division rates, the odds of leukemia diminish.
“With a large population of healthy cells optimized to young, healthy tissue, the ability of mutations, including cancerous mutations, to drive uncontrolled cell proliferation is reduced,” Dr DeGregori said.
However, in old age, tissue decline promotes selection for adaptive mutations, leading to the expansion of potentially oncogenic HSC clones that will again increase the risk of leukemia.
Thus, this research shows that, in early life, leukemias are driven by mutation and drift. And in later life, leukemias are driven by mutation and selection.
“We show that leukemias of children and older adults are different diseases, forged by different evolutionary forces and propagated under different circumstances,” Dr DeGregori said.
He and his colleagues believe this understanding raises the possibility of a new approach to cancer treatment. Perhaps researchers could find a way to manipulate the parameters of cell evolution or manipulate the tissue ecosystem to decrease cancer risk. ![]()
HSPCs shape their own environment, team says

in the bone marrow
New research has revealed a mechanism through which hematopoietic stem and progenitor cells (HSPCs) control both their own proliferation and the characteristics of the niche that houses them.
Researchers detected high expression of the protein E-selectin ligand-1 (ESL-1) in HSPCs and also found that ESL-1 controls HSPCs’ production of the cytokine TGF-β.
The team said this is important because TGF-β has antiproliferative properties and is essential for impeding the loss of HSPCs in some diseases, such as some types of anemia.
The researchers also showed that HSPCs lacking ESL-1 are resistant to chemotherapeutic and cytotoxic agents.
These results suggest ESL-1 is a potential target for therapies aimed at improving bone marrow regeneration after chemotherapy or for expanding the HSPC population in preparation for donation.
Magdalena Leiva, PhD, of Centro Nacional de Investigaciones Cardiovasculares in Madrid, Spain, and her colleagues reported these findings in Nature Communications.
The researchers first found that ESL-1 deficiency causes HSPC quiescence and expansion, and elevated TGF-β causes quiescence in the absence of ESL-1. In addition, ESL-1 controls HSPC proliferation independently of E-selectin, and HSPCs are a relevant source of TGF-β.
The team also discovered that ESL-1 exerts local effects on distinct cell populations in the stromal niche. They found that hematopoietic-borne ESL-1 can control HSPC proliferation directly through cytokine secretion, and/or indirectly through repressive effects on supportive niche cells.
According to Dr Leiva, this finding opens the path to new therapies “that use genetically modified stem cells to treat hematological diseases, such as certain types of leukemia, in which the hematopoietic niche and HSPCs are very affected.”
The researchers made these discoveries by analyzing the bone marrow of mice deficient in ESL-1. In the absence of ESL-1, HSPCs proliferated less and were therefore of superior quality and more suitable for potential therapeutic applications, the team found.
“We see that these cells are resistant to processes associated with bone marrow damage, such as cell death triggered by cytotoxic agents,” Dr Leiva said.
She and her colleagues found that stem cells lacking ESL-1 were resistant to the deleterious effects of 5-fluorouracil and hydroxyurea. They said this suggests ESL-1 is a possible therapeutic target for improved regeneration of the bone marrow during chemotherapy. ![]()

in the bone marrow
New research has revealed a mechanism through which hematopoietic stem and progenitor cells (HSPCs) control both their own proliferation and the characteristics of the niche that houses them.
Researchers detected high expression of the protein E-selectin ligand-1 (ESL-1) in HSPCs and also found that ESL-1 controls HSPCs’ production of the cytokine TGF-β.
The team said this is important because TGF-β has antiproliferative properties and is essential for impeding the loss of HSPCs in some diseases, such as some types of anemia.
The researchers also showed that HSPCs lacking ESL-1 are resistant to chemotherapeutic and cytotoxic agents.
These results suggest ESL-1 is a potential target for therapies aimed at improving bone marrow regeneration after chemotherapy or for expanding the HSPC population in preparation for donation.
Magdalena Leiva, PhD, of Centro Nacional de Investigaciones Cardiovasculares in Madrid, Spain, and her colleagues reported these findings in Nature Communications.
The researchers first found that ESL-1 deficiency causes HSPC quiescence and expansion, and elevated TGF-β causes quiescence in the absence of ESL-1. In addition, ESL-1 controls HSPC proliferation independently of E-selectin, and HSPCs are a relevant source of TGF-β.
The team also discovered that ESL-1 exerts local effects on distinct cell populations in the stromal niche. They found that hematopoietic-borne ESL-1 can control HSPC proliferation directly through cytokine secretion, and/or indirectly through repressive effects on supportive niche cells.
According to Dr Leiva, this finding opens the path to new therapies “that use genetically modified stem cells to treat hematological diseases, such as certain types of leukemia, in which the hematopoietic niche and HSPCs are very affected.”
The researchers made these discoveries by analyzing the bone marrow of mice deficient in ESL-1. In the absence of ESL-1, HSPCs proliferated less and were therefore of superior quality and more suitable for potential therapeutic applications, the team found.
“We see that these cells are resistant to processes associated with bone marrow damage, such as cell death triggered by cytotoxic agents,” Dr Leiva said.
She and her colleagues found that stem cells lacking ESL-1 were resistant to the deleterious effects of 5-fluorouracil and hydroxyurea. They said this suggests ESL-1 is a possible therapeutic target for improved regeneration of the bone marrow during chemotherapy. ![]()

in the bone marrow
New research has revealed a mechanism through which hematopoietic stem and progenitor cells (HSPCs) control both their own proliferation and the characteristics of the niche that houses them.
Researchers detected high expression of the protein E-selectin ligand-1 (ESL-1) in HSPCs and also found that ESL-1 controls HSPCs’ production of the cytokine TGF-β.
The team said this is important because TGF-β has antiproliferative properties and is essential for impeding the loss of HSPCs in some diseases, such as some types of anemia.
The researchers also showed that HSPCs lacking ESL-1 are resistant to chemotherapeutic and cytotoxic agents.
These results suggest ESL-1 is a potential target for therapies aimed at improving bone marrow regeneration after chemotherapy or for expanding the HSPC population in preparation for donation.
Magdalena Leiva, PhD, of Centro Nacional de Investigaciones Cardiovasculares in Madrid, Spain, and her colleagues reported these findings in Nature Communications.
The researchers first found that ESL-1 deficiency causes HSPC quiescence and expansion, and elevated TGF-β causes quiescence in the absence of ESL-1. In addition, ESL-1 controls HSPC proliferation independently of E-selectin, and HSPCs are a relevant source of TGF-β.
The team also discovered that ESL-1 exerts local effects on distinct cell populations in the stromal niche. They found that hematopoietic-borne ESL-1 can control HSPC proliferation directly through cytokine secretion, and/or indirectly through repressive effects on supportive niche cells.
According to Dr Leiva, this finding opens the path to new therapies “that use genetically modified stem cells to treat hematological diseases, such as certain types of leukemia, in which the hematopoietic niche and HSPCs are very affected.”
The researchers made these discoveries by analyzing the bone marrow of mice deficient in ESL-1. In the absence of ESL-1, HSPCs proliferated less and were therefore of superior quality and more suitable for potential therapeutic applications, the team found.
“We see that these cells are resistant to processes associated with bone marrow damage, such as cell death triggered by cytotoxic agents,” Dr Leiva said.
She and her colleagues found that stem cells lacking ESL-1 were resistant to the deleterious effects of 5-fluorouracil and hydroxyurea. They said this suggests ESL-1 is a possible therapeutic target for improved regeneration of the bone marrow during chemotherapy. ![]()
Increasing eligibility for engineered T-cell therapy
Image courtesy of NIAID
A new study suggests that having a certain type of cancer or receiving certain chemotherapeutic agents
can affect T-cell function and make patients ineligible for engineered T-cell therapy.
However, researchers found that proper timing of T-cell collection can increase the number of patients eiligible for the therapy.
And the team developed a culture technique that can boost T cells’ fitness for expansion, which can increase eligibility as well.
Nathan Singh, MD, of the University of Pennsylvania in Philadelphia, and his colleagues described this work in Science Translational Medicine.
The researchers set out to determine why some patients’ T cells fail to multiply in culture. The team studied T cells from children with acute lymphoblastic leukemia (ALL) or non-Hodgkin lymphoma (NHL) who were undergoing chemotherapy. (NHL subtypes included Burkitt lymphoma, diffuse large B-cell lymphoma, primary mediastinal large B-cell lymphoma, primary lymphoma of bone, and follicular lymphoma.)
The researchers found that T cells from patients with ALL expanded better in culture than those from patients with NHL.
The team said a threshold of greater than 5-fold expansion during test expansion was associated with a high likelihood of successful clinical expansion.
Nearly 80% of patients with ALL met this threshold at diagnosis, but the rate declined over the course of therapy, falling to about 40% during maintenance therapy.
About 25% of NHL patients met the threshold at diagnosis, but few samples demonstrated any expansion after therapy began (12.5% of samples at all remaining time points tested).
The researchers said the difference in the proportion of ALL and NHL samples that met the expansion threshold was significant at all time points tested.
Analysis revealed that ALL patients had higher numbers of naïve T cells and stem central memory T cells, T cell subtypes known to be highly potent and proliferative with an enhanced capacity for self-renewal.
The researchers also found that certain chemotherapy drugs—namely, cyclophosphamide and cytarabine—selectively depleted early lineage T cells.
Fortunately, the team discovered that poor expansion can be rescued by exposing T cells to signaling molecules that stimulate T-cell activity. Culture with IL-7 and IL-15 boosted the expansion capacity of T cells from patients with NHL and those with ALL.
The researchers therefore concluded that using this culture technique or collecting T cells prior to chemotherapy can increase the number of patients eligible for engineered T-cell therapy. ![]()

Image courtesy of NIAID
A new study suggests that having a certain type of cancer or receiving certain chemotherapeutic agents
can affect T-cell function and make patients ineligible for engineered T-cell therapy.
However, researchers found that proper timing of T-cell collection can increase the number of patients eiligible for the therapy.
And the team developed a culture technique that can boost T cells’ fitness for expansion, which can increase eligibility as well.
Nathan Singh, MD, of the University of Pennsylvania in Philadelphia, and his colleagues described this work in Science Translational Medicine.
The researchers set out to determine why some patients’ T cells fail to multiply in culture. The team studied T cells from children with acute lymphoblastic leukemia (ALL) or non-Hodgkin lymphoma (NHL) who were undergoing chemotherapy. (NHL subtypes included Burkitt lymphoma, diffuse large B-cell lymphoma, primary mediastinal large B-cell lymphoma, primary lymphoma of bone, and follicular lymphoma.)
The researchers found that T cells from patients with ALL expanded better in culture than those from patients with NHL.
The team said a threshold of greater than 5-fold expansion during test expansion was associated with a high likelihood of successful clinical expansion.
Nearly 80% of patients with ALL met this threshold at diagnosis, but the rate declined over the course of therapy, falling to about 40% during maintenance therapy.
About 25% of NHL patients met the threshold at diagnosis, but few samples demonstrated any expansion after therapy began (12.5% of samples at all remaining time points tested).
The researchers said the difference in the proportion of ALL and NHL samples that met the expansion threshold was significant at all time points tested.
Analysis revealed that ALL patients had higher numbers of naïve T cells and stem central memory T cells, T cell subtypes known to be highly potent and proliferative with an enhanced capacity for self-renewal.
The researchers also found that certain chemotherapy drugs—namely, cyclophosphamide and cytarabine—selectively depleted early lineage T cells.
Fortunately, the team discovered that poor expansion can be rescued by exposing T cells to signaling molecules that stimulate T-cell activity. Culture with IL-7 and IL-15 boosted the expansion capacity of T cells from patients with NHL and those with ALL.
The researchers therefore concluded that using this culture technique or collecting T cells prior to chemotherapy can increase the number of patients eligible for engineered T-cell therapy. ![]()

Image courtesy of NIAID
A new study suggests that having a certain type of cancer or receiving certain chemotherapeutic agents
can affect T-cell function and make patients ineligible for engineered T-cell therapy.
However, researchers found that proper timing of T-cell collection can increase the number of patients eiligible for the therapy.
And the team developed a culture technique that can boost T cells’ fitness for expansion, which can increase eligibility as well.
Nathan Singh, MD, of the University of Pennsylvania in Philadelphia, and his colleagues described this work in Science Translational Medicine.
The researchers set out to determine why some patients’ T cells fail to multiply in culture. The team studied T cells from children with acute lymphoblastic leukemia (ALL) or non-Hodgkin lymphoma (NHL) who were undergoing chemotherapy. (NHL subtypes included Burkitt lymphoma, diffuse large B-cell lymphoma, primary mediastinal large B-cell lymphoma, primary lymphoma of bone, and follicular lymphoma.)
The researchers found that T cells from patients with ALL expanded better in culture than those from patients with NHL.
The team said a threshold of greater than 5-fold expansion during test expansion was associated with a high likelihood of successful clinical expansion.
Nearly 80% of patients with ALL met this threshold at diagnosis, but the rate declined over the course of therapy, falling to about 40% during maintenance therapy.
About 25% of NHL patients met the threshold at diagnosis, but few samples demonstrated any expansion after therapy began (12.5% of samples at all remaining time points tested).
The researchers said the difference in the proportion of ALL and NHL samples that met the expansion threshold was significant at all time points tested.
Analysis revealed that ALL patients had higher numbers of naïve T cells and stem central memory T cells, T cell subtypes known to be highly potent and proliferative with an enhanced capacity for self-renewal.
The researchers also found that certain chemotherapy drugs—namely, cyclophosphamide and cytarabine—selectively depleted early lineage T cells.
Fortunately, the team discovered that poor expansion can be rescued by exposing T cells to signaling molecules that stimulate T-cell activity. Culture with IL-7 and IL-15 boosted the expansion capacity of T cells from patients with NHL and those with ALL.
The researchers therefore concluded that using this culture technique or collecting T cells prior to chemotherapy can increase the number of patients eligible for engineered T-cell therapy. ![]()
US cancer stats: The good and the bad

patient and her father
Photo by Rhoda Baer
The American Cancer Society’s 2016 report on cancer in the US suggests that, in recent years, overall trends in cancer incidence have remained stable for women and declined for men.
However, the rates of certain malignancies are on the rise. This includes some leukemia subtypes for men and women, as well as myeloma for men.
Leukemia is the leading cause of cancer death for men ages 20 to 39, but leukemia is no longer the leading cause of cancer death among children and adolescents (of both genders).
These and other data are included in the report, which is published in CA: A Cancer Journal for Clinicians.
The report estimates there will be 1,685,210 new cancer cases and 595,690 cancer deaths in the US in 2016. This includes 81,080 new lymphoma cases and 21,270 lymphoma deaths, 60,140 new leukemia cases and 24,400 leukemia deaths, and 30,330 new myeloma cases and 12,650 myeloma deaths.
Cancer incidence over time
The report suggests the overall cancer incidence for women has been stable from 1998 to 2012. But the incidence for men has declined by 3.1% per year from 2009 to 2012, with one-half of the drop in men due to recent rapid declines in prostate cancer diagnoses as prostate-specific antigen testing decreases.
Incidence rates increased from 2003 to 2012 among both men and women for some leukemia subtypes and for cancers of the tongue, tonsil, small intestine, liver, pancreas, kidney, renal pelvis, and thyroid.
Incidence rates increased in men for melanoma, myeloma, and cancers of the breast, testis, and oropharynx. Among women, incidence rates increased for cancers of the anus, vulva, and uterine corpus.
Cancer deaths
The rate of cancer deaths in the US has dropped 23% from its peak in 1991 to 2012. The incidence of cancer death was 215.1 per 100,000 in 1991 and 166.4 per 100,000 in 2012.
The decline is larger in men (28% since 1990) than in women (19% since 1991). Over the past decade of data, the rate dropped by 1.8% per year in men and 1.4% per year in women.
The decline in cancer death rates over the past 2 decades is driven by continued decreases in death rates for the 4 major cancer sites: lung, breast, prostate, and colon/rectum.
Breast cancer is the leading cause of cancer death in women ages 20 to 59, while lung cancer is the leading cause of cancer death in women 60 and older.
Among men, leukemia is the leading cause of cancer death for those ages 20 to 39, whereas lung cancer ranks first among men 40 and older.
Among children and adolescents (0-19), brain cancer has surpassed leukemia as the leading cause of cancer death, a result of more rapid therapeutic advances against leukemia.
The report also features an analysis of leading causes of death by state. It shows that, even as cancer remains the second leading cause of death nationwide, steep drops in deaths from heart disease have made cancer the leading cause of death in 21 states: Alaska, Arizona, Colorado, Delaware, Florida, Georgia, Idaho, Kansas, Maine, Massachusetts, Minnesota, Montana, Nebraska, New Hampshire, New Mexico, North Carolina, Oregon, South Carolina, Vermont, Virginia, and Washington.
In addition, cancer is the leading cause of death among adults ages 40 to 79 and among both Hispanics and Asian/Pacific Islanders, who together make up one-quarter of the US population.
Heart disease remains the top cause of death overall in the US. In 2012, there were 599,711 (24%) deaths from heart disease, compared to 582,623 (23%) deaths from cancer.
“We’re gratified to see cancer death rates continuing to drop,” said Otis W. Brawley, MD, chief medical officer of the American Cancer Society.
“But the fact that cancer is nonetheless becoming the top cause of death in many populations is a strong reminder that the fight is not over.” ![]()

patient and her father
Photo by Rhoda Baer
The American Cancer Society’s 2016 report on cancer in the US suggests that, in recent years, overall trends in cancer incidence have remained stable for women and declined for men.
However, the rates of certain malignancies are on the rise. This includes some leukemia subtypes for men and women, as well as myeloma for men.
Leukemia is the leading cause of cancer death for men ages 20 to 39, but leukemia is no longer the leading cause of cancer death among children and adolescents (of both genders).
These and other data are included in the report, which is published in CA: A Cancer Journal for Clinicians.
The report estimates there will be 1,685,210 new cancer cases and 595,690 cancer deaths in the US in 2016. This includes 81,080 new lymphoma cases and 21,270 lymphoma deaths, 60,140 new leukemia cases and 24,400 leukemia deaths, and 30,330 new myeloma cases and 12,650 myeloma deaths.
Cancer incidence over time
The report suggests the overall cancer incidence for women has been stable from 1998 to 2012. But the incidence for men has declined by 3.1% per year from 2009 to 2012, with one-half of the drop in men due to recent rapid declines in prostate cancer diagnoses as prostate-specific antigen testing decreases.
Incidence rates increased from 2003 to 2012 among both men and women for some leukemia subtypes and for cancers of the tongue, tonsil, small intestine, liver, pancreas, kidney, renal pelvis, and thyroid.
Incidence rates increased in men for melanoma, myeloma, and cancers of the breast, testis, and oropharynx. Among women, incidence rates increased for cancers of the anus, vulva, and uterine corpus.
Cancer deaths
The rate of cancer deaths in the US has dropped 23% from its peak in 1991 to 2012. The incidence of cancer death was 215.1 per 100,000 in 1991 and 166.4 per 100,000 in 2012.
The decline is larger in men (28% since 1990) than in women (19% since 1991). Over the past decade of data, the rate dropped by 1.8% per year in men and 1.4% per year in women.
The decline in cancer death rates over the past 2 decades is driven by continued decreases in death rates for the 4 major cancer sites: lung, breast, prostate, and colon/rectum.
Breast cancer is the leading cause of cancer death in women ages 20 to 59, while lung cancer is the leading cause of cancer death in women 60 and older.
Among men, leukemia is the leading cause of cancer death for those ages 20 to 39, whereas lung cancer ranks first among men 40 and older.
Among children and adolescents (0-19), brain cancer has surpassed leukemia as the leading cause of cancer death, a result of more rapid therapeutic advances against leukemia.
The report also features an analysis of leading causes of death by state. It shows that, even as cancer remains the second leading cause of death nationwide, steep drops in deaths from heart disease have made cancer the leading cause of death in 21 states: Alaska, Arizona, Colorado, Delaware, Florida, Georgia, Idaho, Kansas, Maine, Massachusetts, Minnesota, Montana, Nebraska, New Hampshire, New Mexico, North Carolina, Oregon, South Carolina, Vermont, Virginia, and Washington.
In addition, cancer is the leading cause of death among adults ages 40 to 79 and among both Hispanics and Asian/Pacific Islanders, who together make up one-quarter of the US population.
Heart disease remains the top cause of death overall in the US. In 2012, there were 599,711 (24%) deaths from heart disease, compared to 582,623 (23%) deaths from cancer.
“We’re gratified to see cancer death rates continuing to drop,” said Otis W. Brawley, MD, chief medical officer of the American Cancer Society.
“But the fact that cancer is nonetheless becoming the top cause of death in many populations is a strong reminder that the fight is not over.” ![]()

patient and her father
Photo by Rhoda Baer
The American Cancer Society’s 2016 report on cancer in the US suggests that, in recent years, overall trends in cancer incidence have remained stable for women and declined for men.
However, the rates of certain malignancies are on the rise. This includes some leukemia subtypes for men and women, as well as myeloma for men.
Leukemia is the leading cause of cancer death for men ages 20 to 39, but leukemia is no longer the leading cause of cancer death among children and adolescents (of both genders).
These and other data are included in the report, which is published in CA: A Cancer Journal for Clinicians.
The report estimates there will be 1,685,210 new cancer cases and 595,690 cancer deaths in the US in 2016. This includes 81,080 new lymphoma cases and 21,270 lymphoma deaths, 60,140 new leukemia cases and 24,400 leukemia deaths, and 30,330 new myeloma cases and 12,650 myeloma deaths.
Cancer incidence over time
The report suggests the overall cancer incidence for women has been stable from 1998 to 2012. But the incidence for men has declined by 3.1% per year from 2009 to 2012, with one-half of the drop in men due to recent rapid declines in prostate cancer diagnoses as prostate-specific antigen testing decreases.
Incidence rates increased from 2003 to 2012 among both men and women for some leukemia subtypes and for cancers of the tongue, tonsil, small intestine, liver, pancreas, kidney, renal pelvis, and thyroid.
Incidence rates increased in men for melanoma, myeloma, and cancers of the breast, testis, and oropharynx. Among women, incidence rates increased for cancers of the anus, vulva, and uterine corpus.
Cancer deaths
The rate of cancer deaths in the US has dropped 23% from its peak in 1991 to 2012. The incidence of cancer death was 215.1 per 100,000 in 1991 and 166.4 per 100,000 in 2012.
The decline is larger in men (28% since 1990) than in women (19% since 1991). Over the past decade of data, the rate dropped by 1.8% per year in men and 1.4% per year in women.
The decline in cancer death rates over the past 2 decades is driven by continued decreases in death rates for the 4 major cancer sites: lung, breast, prostate, and colon/rectum.
Breast cancer is the leading cause of cancer death in women ages 20 to 59, while lung cancer is the leading cause of cancer death in women 60 and older.
Among men, leukemia is the leading cause of cancer death for those ages 20 to 39, whereas lung cancer ranks first among men 40 and older.
Among children and adolescents (0-19), brain cancer has surpassed leukemia as the leading cause of cancer death, a result of more rapid therapeutic advances against leukemia.
The report also features an analysis of leading causes of death by state. It shows that, even as cancer remains the second leading cause of death nationwide, steep drops in deaths from heart disease have made cancer the leading cause of death in 21 states: Alaska, Arizona, Colorado, Delaware, Florida, Georgia, Idaho, Kansas, Maine, Massachusetts, Minnesota, Montana, Nebraska, New Hampshire, New Mexico, North Carolina, Oregon, South Carolina, Vermont, Virginia, and Washington.
In addition, cancer is the leading cause of death among adults ages 40 to 79 and among both Hispanics and Asian/Pacific Islanders, who together make up one-quarter of the US population.
Heart disease remains the top cause of death overall in the US. In 2012, there were 599,711 (24%) deaths from heart disease, compared to 582,623 (23%) deaths from cancer.
“We’re gratified to see cancer death rates continuing to drop,” said Otis W. Brawley, MD, chief medical officer of the American Cancer Society.
“But the fact that cancer is nonetheless becoming the top cause of death in many populations is a strong reminder that the fight is not over.” ![]()
KIR2DL5B genotype predicts outcome in chronic phase–CML
The presence of KIR2DL5B was associated with lower rates of major molecular response (MMR), transformation-free survival, and event-free survival (but not overall survival) in patients with chronic phase–chronic myeloid leukemia (CP-CML) treated with sequential imatinib/nilotinib, according to researchers.
Univariate analysis demonstrated a significant association between KIR2DL5B and achievement of a major molecular response, with hazard ratio 0.423 (95% CI, 0.262-0.682; P less than .001). Other KIR genotypes, KIR2DL2pos and KIR2DS3pos, were also associated with inferior achievement of MMR, probably because of their association with KIR2DL5B due to linkage disequilibrium among KIR genes, according to the investigators.
“Our findings suggest that even with the potent second-generation TKI [tyrosine kinase inhibitor] nilotinib, KIR genotypes, a predetermined genetic host factor, may still be one of the most discriminatory prognostic markers available at baseline,” wrote Dr. David T. Yeung of the department of genetics and molecular pathology, Centre for Cancer Biology and the University of Adelaide, South Australia, and colleagues (Blood 2015 Dec 17. doi:10.1182/blood-2015-07-655589).
Killer immunoglobulin-like receptors (KIRs) contribute to natural killer (NK) cell–mediated killing of tumor cells, in both activating and inhibitory roles. Normal cells are spared through actions of inhibitory KIRs. Although the mechanism underlying the association between KIR2DL5B and CP-CML treatment outcomes is still unclear, the gene encodes an inhibitory KIR receptor, the absence of which may increase efficiency of NK-mediated killing of leukemic stem cells, researchers suggested.
The Therapeutic Intensification in De Novo Leukaemia (TIDEL-II) study included 210 patients with CP-CML who were treated with imatinib initially, and nilotinib subsequently if predetermined molecular targets were not met. The KIR substudy included 148 patients with samples available for genotyping.
KIR genotype frequencies observed in this study were similar to other white populations reported in the Allele Frequency Database.
Early molecular response was also significantly associated with treatment outcomes, independent of KIR prognostic significance, and may add additional prognostic information, available 3 months after treatment commences.
“In contrast, KIR2DL5B can identify, at baseline, the 20% of patients with a transformation risk of [about] 10% over 2 years versus the 80% of patients with a transformation risk of less than 3%,” the authors wrote. They suggest that KIR2DL5B, combined with other predictive markers, may enable targeted early interventions to improve outcomes.
The presence of KIR2DL5B was associated with lower rates of major molecular response (MMR), transformation-free survival, and event-free survival (but not overall survival) in patients with chronic phase–chronic myeloid leukemia (CP-CML) treated with sequential imatinib/nilotinib, according to researchers.
Univariate analysis demonstrated a significant association between KIR2DL5B and achievement of a major molecular response, with hazard ratio 0.423 (95% CI, 0.262-0.682; P less than .001). Other KIR genotypes, KIR2DL2pos and KIR2DS3pos, were also associated with inferior achievement of MMR, probably because of their association with KIR2DL5B due to linkage disequilibrium among KIR genes, according to the investigators.
“Our findings suggest that even with the potent second-generation TKI [tyrosine kinase inhibitor] nilotinib, KIR genotypes, a predetermined genetic host factor, may still be one of the most discriminatory prognostic markers available at baseline,” wrote Dr. David T. Yeung of the department of genetics and molecular pathology, Centre for Cancer Biology and the University of Adelaide, South Australia, and colleagues (Blood 2015 Dec 17. doi:10.1182/blood-2015-07-655589).
Killer immunoglobulin-like receptors (KIRs) contribute to natural killer (NK) cell–mediated killing of tumor cells, in both activating and inhibitory roles. Normal cells are spared through actions of inhibitory KIRs. Although the mechanism underlying the association between KIR2DL5B and CP-CML treatment outcomes is still unclear, the gene encodes an inhibitory KIR receptor, the absence of which may increase efficiency of NK-mediated killing of leukemic stem cells, researchers suggested.
The Therapeutic Intensification in De Novo Leukaemia (TIDEL-II) study included 210 patients with CP-CML who were treated with imatinib initially, and nilotinib subsequently if predetermined molecular targets were not met. The KIR substudy included 148 patients with samples available for genotyping.
KIR genotype frequencies observed in this study were similar to other white populations reported in the Allele Frequency Database.
Early molecular response was also significantly associated with treatment outcomes, independent of KIR prognostic significance, and may add additional prognostic information, available 3 months after treatment commences.
“In contrast, KIR2DL5B can identify, at baseline, the 20% of patients with a transformation risk of [about] 10% over 2 years versus the 80% of patients with a transformation risk of less than 3%,” the authors wrote. They suggest that KIR2DL5B, combined with other predictive markers, may enable targeted early interventions to improve outcomes.
The presence of KIR2DL5B was associated with lower rates of major molecular response (MMR), transformation-free survival, and event-free survival (but not overall survival) in patients with chronic phase–chronic myeloid leukemia (CP-CML) treated with sequential imatinib/nilotinib, according to researchers.
Univariate analysis demonstrated a significant association between KIR2DL5B and achievement of a major molecular response, with hazard ratio 0.423 (95% CI, 0.262-0.682; P less than .001). Other KIR genotypes, KIR2DL2pos and KIR2DS3pos, were also associated with inferior achievement of MMR, probably because of their association with KIR2DL5B due to linkage disequilibrium among KIR genes, according to the investigators.
“Our findings suggest that even with the potent second-generation TKI [tyrosine kinase inhibitor] nilotinib, KIR genotypes, a predetermined genetic host factor, may still be one of the most discriminatory prognostic markers available at baseline,” wrote Dr. David T. Yeung of the department of genetics and molecular pathology, Centre for Cancer Biology and the University of Adelaide, South Australia, and colleagues (Blood 2015 Dec 17. doi:10.1182/blood-2015-07-655589).
Killer immunoglobulin-like receptors (KIRs) contribute to natural killer (NK) cell–mediated killing of tumor cells, in both activating and inhibitory roles. Normal cells are spared through actions of inhibitory KIRs. Although the mechanism underlying the association between KIR2DL5B and CP-CML treatment outcomes is still unclear, the gene encodes an inhibitory KIR receptor, the absence of which may increase efficiency of NK-mediated killing of leukemic stem cells, researchers suggested.
The Therapeutic Intensification in De Novo Leukaemia (TIDEL-II) study included 210 patients with CP-CML who were treated with imatinib initially, and nilotinib subsequently if predetermined molecular targets were not met. The KIR substudy included 148 patients with samples available for genotyping.
KIR genotype frequencies observed in this study were similar to other white populations reported in the Allele Frequency Database.
Early molecular response was also significantly associated with treatment outcomes, independent of KIR prognostic significance, and may add additional prognostic information, available 3 months after treatment commences.
“In contrast, KIR2DL5B can identify, at baseline, the 20% of patients with a transformation risk of [about] 10% over 2 years versus the 80% of patients with a transformation risk of less than 3%,” the authors wrote. They suggest that KIR2DL5B, combined with other predictive markers, may enable targeted early interventions to improve outcomes.
FROM BLOOD
Key clinical point: The presence of KIR2DL5B was associated with worse outcomes in patients with chronic phase–chronic myeloid leukemia treated with sequential imatinib/nilotinib.
Major finding: Achievement of a major molecular response was associated with the KIR2DL5B genotype (HR, 0.423; 95% CI, 0.262-0.682; P less than .001).
Data source: A substudy of the Therapeutic Intensification in De Novo Leukaemia (TIDEL-II) study that included 148 patients with KIR genotype data available.
Disclosures: Support for the study was provided in part by Novartis. Dr. Yeung reported consulting or advisory roles with Novartis, BMS, and Ariad. Several coauthors reported ties to industry.
Study links leukemia to low UVB exposure

People residing at higher latitudes, with lower exposure to sunlight/ultraviolet B (UVB) rays, have at least a 2-fold greater risk of developing leukemia than equatorial populations, according to research published in PLOS ONE.
“These results suggest that much of the burden of leukemia worldwide is due to the epidemic of vitamin D deficiency we are experiencing in winter in populations distant from the equator,” said Cedric Garland, DrPH, of the University of California San Diego in La Jolla, California.
“People who live in areas with low solar ultraviolet B exposure tend to have low levels of vitamin D metabolites in their blood. These low levels place them at high risk of certain cancers, including leukemia.”
Dr Garland and his colleagues analyzed age-adjusted incidence rates of leukemia in 172 countries and compared that information with cloud cover data from the International Satellite Cloud Climatology Project.
The team found that leukemia rates were highest in countries relatively closer to the poles, such as Australia, New Zealand, Chile, Ireland, Canada, and the United States.
And leukemia rates were lowest in countries closer to the equator, such as Bolivia, Samoa, Madagascar, and Nigeria.
The researchers also discovered that leukemia incidence was inversely associated with cloud-adjusted UVB irradiance in males (P≤0.01) and females (P≤0.01) in both hemispheres.
The association persisted in males (P≤0.05) and females (P≤0.01) after the team controlled for elevation and life expectancy.
The researchers said it’s plausible that the association is due to vitamin D deficiency.
This study follows similar investigations by Dr Garland and his colleagues in which they looked at other cancers, including breast, colon, pancreas, bladder, and multiple myeloma. In each study, the team found that reduced UVB radiation exposure and lower vitamin D levels were associated with higher risks of cancer.
“These studies do not necessarily provide final evidence,” Dr Garland said, “but they have been helpful in the past in identifying associations that have helped minimize cancer risk.” ![]()

People residing at higher latitudes, with lower exposure to sunlight/ultraviolet B (UVB) rays, have at least a 2-fold greater risk of developing leukemia than equatorial populations, according to research published in PLOS ONE.
“These results suggest that much of the burden of leukemia worldwide is due to the epidemic of vitamin D deficiency we are experiencing in winter in populations distant from the equator,” said Cedric Garland, DrPH, of the University of California San Diego in La Jolla, California.
“People who live in areas with low solar ultraviolet B exposure tend to have low levels of vitamin D metabolites in their blood. These low levels place them at high risk of certain cancers, including leukemia.”
Dr Garland and his colleagues analyzed age-adjusted incidence rates of leukemia in 172 countries and compared that information with cloud cover data from the International Satellite Cloud Climatology Project.
The team found that leukemia rates were highest in countries relatively closer to the poles, such as Australia, New Zealand, Chile, Ireland, Canada, and the United States.
And leukemia rates were lowest in countries closer to the equator, such as Bolivia, Samoa, Madagascar, and Nigeria.
The researchers also discovered that leukemia incidence was inversely associated with cloud-adjusted UVB irradiance in males (P≤0.01) and females (P≤0.01) in both hemispheres.
The association persisted in males (P≤0.05) and females (P≤0.01) after the team controlled for elevation and life expectancy.
The researchers said it’s plausible that the association is due to vitamin D deficiency.
This study follows similar investigations by Dr Garland and his colleagues in which they looked at other cancers, including breast, colon, pancreas, bladder, and multiple myeloma. In each study, the team found that reduced UVB radiation exposure and lower vitamin D levels were associated with higher risks of cancer.
“These studies do not necessarily provide final evidence,” Dr Garland said, “but they have been helpful in the past in identifying associations that have helped minimize cancer risk.” ![]()

People residing at higher latitudes, with lower exposure to sunlight/ultraviolet B (UVB) rays, have at least a 2-fold greater risk of developing leukemia than equatorial populations, according to research published in PLOS ONE.
“These results suggest that much of the burden of leukemia worldwide is due to the epidemic of vitamin D deficiency we are experiencing in winter in populations distant from the equator,” said Cedric Garland, DrPH, of the University of California San Diego in La Jolla, California.
“People who live in areas with low solar ultraviolet B exposure tend to have low levels of vitamin D metabolites in their blood. These low levels place them at high risk of certain cancers, including leukemia.”
Dr Garland and his colleagues analyzed age-adjusted incidence rates of leukemia in 172 countries and compared that information with cloud cover data from the International Satellite Cloud Climatology Project.
The team found that leukemia rates were highest in countries relatively closer to the poles, such as Australia, New Zealand, Chile, Ireland, Canada, and the United States.
And leukemia rates were lowest in countries closer to the equator, such as Bolivia, Samoa, Madagascar, and Nigeria.
The researchers also discovered that leukemia incidence was inversely associated with cloud-adjusted UVB irradiance in males (P≤0.01) and females (P≤0.01) in both hemispheres.
The association persisted in males (P≤0.05) and females (P≤0.01) after the team controlled for elevation and life expectancy.
The researchers said it’s plausible that the association is due to vitamin D deficiency.
This study follows similar investigations by Dr Garland and his colleagues in which they looked at other cancers, including breast, colon, pancreas, bladder, and multiple myeloma. In each study, the team found that reduced UVB radiation exposure and lower vitamin D levels were associated with higher risks of cancer.
“These studies do not necessarily provide final evidence,” Dr Garland said, “but they have been helpful in the past in identifying associations that have helped minimize cancer risk.” ![]()
Cardiac abnormalities among childhood cancer survivors

Photo by Bill Branson
A new study has provided additional insight into the development of cardiac abnormalities in adult survivors of childhood cancer.
Researchers analyzed more than 1800 cancer survivors who were exposed to cardiotoxic therapies as children.
The team said they found evidence of cardiac abnormalities in a substantial number of these subjects, many of whom were younger and did not exhibit symptoms of abnormalities.
Daniel A. Mulrooney, MD, of St. Jude Children’s Research Hospital in Memphis, Tennessee, and his colleagues reported these findings in Annals of Internal Medicine.
The team assessed cardiac outcomes among 1853 subjects who were 18 and older and had received cancer-related cardiotoxic therapy at least 10 years earlier.
The subjects were pretty evenly split along gender lines (52.3% male), their median age at cancer diagnosis was 8 (range, 0 to 24), and their median age at evaluation was 31 (range, 18 to 60).
At evaluation, 7.4% of subjects had cardiomyopathy (newly identified in 4.7%), 3.8% had coronary artery disease (newly identified in 2.2%), 28% had valvular regurgitation or stenosis (newly identified in 24.8%), and 4.4% had conduction or rhythm abnormalities (newly identified in 1.4%). All but 5 subjects were asymptomatic.
Multivariable analysis suggested the odds of developing cardiomyopathy were significantly associated with being male (odds ratio [OR]=1.9), receiving anthracycline doses of 250 mg/m2 or greater (OR=2.7), having cardiac radiation exposure greater than 1500 cGy (OR=1.9), and having hypertension (OR=3.0).
Being younger at diagnosis was associated with higher odds of valvular disease. The ORs were 1.5 for patients who were 0 to 4 years of age at diagnosis and 1.3 for patients who were 5 to 9 at diagnosis.
Receiving higher radiation doses was associated with higher odds of valvular disease as well. But associations between radiation and valvular disease varied according to a patient’s anthracycline exposure (interaction P<0.001). The highest odds were among survivors with the highest doses of radiation exposure and any anthracycline exposure (OR=4.5).
The researchers also noted a reduction in the OR for valvular disease among obese patients (OR=0.4) and those with dyslipidemia (OR=0.7).
The team said there were not enough cases of coronary artery disease and conduction or rhythm abnormalities to support a fully adjusted multivariable model. However, it seemed these outcomes were more common with older age (≥40 years) and among patients with cardiac radiation doses of 1500 cGy or greater.
The researchers said this study revealed “considerable cardiovascular disease” in a large cohort of adult survivors of childhood cancer, which suggests a substantial future healthcare burden.
The team believes their findings could guide stratification of risk factors, screening practices, health counseling, and potential therapeutic measures aimed at changing the disease trajectory in this young adult population. ![]()

Photo by Bill Branson
A new study has provided additional insight into the development of cardiac abnormalities in adult survivors of childhood cancer.
Researchers analyzed more than 1800 cancer survivors who were exposed to cardiotoxic therapies as children.
The team said they found evidence of cardiac abnormalities in a substantial number of these subjects, many of whom were younger and did not exhibit symptoms of abnormalities.
Daniel A. Mulrooney, MD, of St. Jude Children’s Research Hospital in Memphis, Tennessee, and his colleagues reported these findings in Annals of Internal Medicine.
The team assessed cardiac outcomes among 1853 subjects who were 18 and older and had received cancer-related cardiotoxic therapy at least 10 years earlier.
The subjects were pretty evenly split along gender lines (52.3% male), their median age at cancer diagnosis was 8 (range, 0 to 24), and their median age at evaluation was 31 (range, 18 to 60).
At evaluation, 7.4% of subjects had cardiomyopathy (newly identified in 4.7%), 3.8% had coronary artery disease (newly identified in 2.2%), 28% had valvular regurgitation or stenosis (newly identified in 24.8%), and 4.4% had conduction or rhythm abnormalities (newly identified in 1.4%). All but 5 subjects were asymptomatic.
Multivariable analysis suggested the odds of developing cardiomyopathy were significantly associated with being male (odds ratio [OR]=1.9), receiving anthracycline doses of 250 mg/m2 or greater (OR=2.7), having cardiac radiation exposure greater than 1500 cGy (OR=1.9), and having hypertension (OR=3.0).
Being younger at diagnosis was associated with higher odds of valvular disease. The ORs were 1.5 for patients who were 0 to 4 years of age at diagnosis and 1.3 for patients who were 5 to 9 at diagnosis.
Receiving higher radiation doses was associated with higher odds of valvular disease as well. But associations between radiation and valvular disease varied according to a patient’s anthracycline exposure (interaction P<0.001). The highest odds were among survivors with the highest doses of radiation exposure and any anthracycline exposure (OR=4.5).
The researchers also noted a reduction in the OR for valvular disease among obese patients (OR=0.4) and those with dyslipidemia (OR=0.7).
The team said there were not enough cases of coronary artery disease and conduction or rhythm abnormalities to support a fully adjusted multivariable model. However, it seemed these outcomes were more common with older age (≥40 years) and among patients with cardiac radiation doses of 1500 cGy or greater.
The researchers said this study revealed “considerable cardiovascular disease” in a large cohort of adult survivors of childhood cancer, which suggests a substantial future healthcare burden.
The team believes their findings could guide stratification of risk factors, screening practices, health counseling, and potential therapeutic measures aimed at changing the disease trajectory in this young adult population. ![]()

Photo by Bill Branson
A new study has provided additional insight into the development of cardiac abnormalities in adult survivors of childhood cancer.
Researchers analyzed more than 1800 cancer survivors who were exposed to cardiotoxic therapies as children.
The team said they found evidence of cardiac abnormalities in a substantial number of these subjects, many of whom were younger and did not exhibit symptoms of abnormalities.
Daniel A. Mulrooney, MD, of St. Jude Children’s Research Hospital in Memphis, Tennessee, and his colleagues reported these findings in Annals of Internal Medicine.
The team assessed cardiac outcomes among 1853 subjects who were 18 and older and had received cancer-related cardiotoxic therapy at least 10 years earlier.
The subjects were pretty evenly split along gender lines (52.3% male), their median age at cancer diagnosis was 8 (range, 0 to 24), and their median age at evaluation was 31 (range, 18 to 60).
At evaluation, 7.4% of subjects had cardiomyopathy (newly identified in 4.7%), 3.8% had coronary artery disease (newly identified in 2.2%), 28% had valvular regurgitation or stenosis (newly identified in 24.8%), and 4.4% had conduction or rhythm abnormalities (newly identified in 1.4%). All but 5 subjects were asymptomatic.
Multivariable analysis suggested the odds of developing cardiomyopathy were significantly associated with being male (odds ratio [OR]=1.9), receiving anthracycline doses of 250 mg/m2 or greater (OR=2.7), having cardiac radiation exposure greater than 1500 cGy (OR=1.9), and having hypertension (OR=3.0).
Being younger at diagnosis was associated with higher odds of valvular disease. The ORs were 1.5 for patients who were 0 to 4 years of age at diagnosis and 1.3 for patients who were 5 to 9 at diagnosis.
Receiving higher radiation doses was associated with higher odds of valvular disease as well. But associations between radiation and valvular disease varied according to a patient’s anthracycline exposure (interaction P<0.001). The highest odds were among survivors with the highest doses of radiation exposure and any anthracycline exposure (OR=4.5).
The researchers also noted a reduction in the OR for valvular disease among obese patients (OR=0.4) and those with dyslipidemia (OR=0.7).
The team said there were not enough cases of coronary artery disease and conduction or rhythm abnormalities to support a fully adjusted multivariable model. However, it seemed these outcomes were more common with older age (≥40 years) and among patients with cardiac radiation doses of 1500 cGy or greater.
The researchers said this study revealed “considerable cardiovascular disease” in a large cohort of adult survivors of childhood cancer, which suggests a substantial future healthcare burden.
The team believes their findings could guide stratification of risk factors, screening practices, health counseling, and potential therapeutic measures aimed at changing the disease trajectory in this young adult population.
Paste may reduce radiation-induced fibrosis
woman for radiation
Photo by Rhoda Baer
A topical paste can reduce fibrosis caused by radiation therapy, according to preclinical research published in The FASEB Journal.
The study addressed a type of fibrosis called radiation dermatitis, in which radiation applied to the skin causes the buildup of fibrotic tissue and skin thickening.
To test their topical paste, researchers mimicked the development of radiation dermatitis in mice.
They exposed the mice’s skin to a single dose of 40 Gy, an amount of radiation similar to what patients undergoing anticancer radiation typically receive over 5 weeks.
Some of the irradiated animals were wild-type mice, while others were genetically engineered to lack the A2A receptor (A2AR). The researchers had previously shown that occupancy of A2AR induces collagen production.
The wild-type mice went on to receive placebo or daily treatment with ZM241385, a paste made with the research team’s patented A2AR blocker. The paste contains 2.5 milligrams of active ingredient per milliliter of 3% carboxymethyl cellulose, a gum “binder.”
A month after exposure, wild-type mice that received placebo had a nearly 2-fold increase in the amount of collagen and skin thickness. These mice also experienced epithelial hyperplasia.
On the other hand, mice treated with ZM241385 accumulated only 10% more skin-thickening collagen. ZM241385 treatment reduced the number of myofibroblasts, collagen fibrils, proliferating keratinocytes, and angiogenesis when compared to placebo. And the paste prevented epithelial hyperplasia.
Like ZM241385-treated mice, A2AR knockout mice did not have the excessive collagen production and skin thickening observed in placebo-treated wild-type mice. The knockout mice also exhibited reductions in myofibroblast content, angiogenesis, and epithelial hyperplasia.
The researchers noted that radiation-induced changes in the dermis and epidermis were accompanied by an infiltrate of T cells, which was prevented in both ZM241385-treated and A2AR knockout mice.
“Our latest study is the first to demonstrate that blocking or deleting the A2A receptor can be useful in reducing radiation-induced scarring in skin,” said study author Bruce Cronstein, MD, of New York University School of Medicine in New York, New York.
“The study also suggests that adenosine A2A receptor antagonists may have broad applications as drug therapies for preventing fibrosis and scarring, not just in the liver but also in the skin.”
If further experiments prove successful, Dr Cronstein said, clinicians treating early stage cancers with radiation could eventually prescribe an A2AR inhibitor paste to prevent fibrosis. He said his team next plans to study the mechanism underlying A2AR’s role in fibrosis.

woman for radiation
Photo by Rhoda Baer
A topical paste can reduce fibrosis caused by radiation therapy, according to preclinical research published in The FASEB Journal.
The study addressed a type of fibrosis called radiation dermatitis, in which radiation applied to the skin causes the buildup of fibrotic tissue and skin thickening.
To test their topical paste, researchers mimicked the development of radiation dermatitis in mice.
They exposed the mice’s skin to a single dose of 40 Gy, an amount of radiation similar to what patients undergoing anticancer radiation typically receive over 5 weeks.
Some of the irradiated animals were wild-type mice, while others were genetically engineered to lack the A2A receptor (A2AR). The researchers had previously shown that occupancy of A2AR induces collagen production.
The wild-type mice went on to receive placebo or daily treatment with ZM241385, a paste made with the research team’s patented A2AR blocker. The paste contains 2.5 milligrams of active ingredient per milliliter of 3% carboxymethyl cellulose, a gum “binder.”
A month after exposure, wild-type mice that received placebo had a nearly 2-fold increase in the amount of collagen and skin thickness. These mice also experienced epithelial hyperplasia.
On the other hand, mice treated with ZM241385 accumulated only 10% more skin-thickening collagen. ZM241385 treatment reduced the number of myofibroblasts, collagen fibrils, proliferating keratinocytes, and angiogenesis when compared to placebo. And the paste prevented epithelial hyperplasia.
Like ZM241385-treated mice, A2AR knockout mice did not have the excessive collagen production and skin thickening observed in placebo-treated wild-type mice. The knockout mice also exhibited reductions in myofibroblast content, angiogenesis, and epithelial hyperplasia.
The researchers noted that radiation-induced changes in the dermis and epidermis were accompanied by an infiltrate of T cells, which was prevented in both ZM241385-treated and A2AR knockout mice.
“Our latest study is the first to demonstrate that blocking or deleting the A2A receptor can be useful in reducing radiation-induced scarring in skin,” said study author Bruce Cronstein, MD, of New York University School of Medicine in New York, New York.
“The study also suggests that adenosine A2A receptor antagonists may have broad applications as drug therapies for preventing fibrosis and scarring, not just in the liver but also in the skin.”
If further experiments prove successful, Dr Cronstein said, clinicians treating early stage cancers with radiation could eventually prescribe an A2AR inhibitor paste to prevent fibrosis. He said his team next plans to study the mechanism underlying A2AR’s role in fibrosis.

woman for radiation
Photo by Rhoda Baer
A topical paste can reduce fibrosis caused by radiation therapy, according to preclinical research published in The FASEB Journal.
The study addressed a type of fibrosis called radiation dermatitis, in which radiation applied to the skin causes the buildup of fibrotic tissue and skin thickening.
To test their topical paste, researchers mimicked the development of radiation dermatitis in mice.
They exposed the mice’s skin to a single dose of 40 Gy, an amount of radiation similar to what patients undergoing anticancer radiation typically receive over 5 weeks.
Some of the irradiated animals were wild-type mice, while others were genetically engineered to lack the A2A receptor (A2AR). The researchers had previously shown that occupancy of A2AR induces collagen production.
The wild-type mice went on to receive placebo or daily treatment with ZM241385, a paste made with the research team’s patented A2AR blocker. The paste contains 2.5 milligrams of active ingredient per milliliter of 3% carboxymethyl cellulose, a gum “binder.”
A month after exposure, wild-type mice that received placebo had a nearly 2-fold increase in the amount of collagen and skin thickness. These mice also experienced epithelial hyperplasia.
On the other hand, mice treated with ZM241385 accumulated only 10% more skin-thickening collagen. ZM241385 treatment reduced the number of myofibroblasts, collagen fibrils, proliferating keratinocytes, and angiogenesis when compared to placebo. And the paste prevented epithelial hyperplasia.
Like ZM241385-treated mice, A2AR knockout mice did not have the excessive collagen production and skin thickening observed in placebo-treated wild-type mice. The knockout mice also exhibited reductions in myofibroblast content, angiogenesis, and epithelial hyperplasia.
The researchers noted that radiation-induced changes in the dermis and epidermis were accompanied by an infiltrate of T cells, which was prevented in both ZM241385-treated and A2AR knockout mice.
“Our latest study is the first to demonstrate that blocking or deleting the A2A receptor can be useful in reducing radiation-induced scarring in skin,” said study author Bruce Cronstein, MD, of New York University School of Medicine in New York, New York.
“The study also suggests that adenosine A2A receptor antagonists may have broad applications as drug therapies for preventing fibrosis and scarring, not just in the liver but also in the skin.”
If further experiments prove successful, Dr Cronstein said, clinicians treating early stage cancers with radiation could eventually prescribe an A2AR inhibitor paste to prevent fibrosis. He said his team next plans to study the mechanism underlying A2AR’s role in fibrosis.
Cancer drug discovery database goes 3D

Photo by Rhoda Baer
Researchers have updated the canSAR database, a tool designed to aid cancer drug discovery, by adding 3D structures of faulty proteins and maps of cancer’s communication networks.
The canSAR database brings together biological, chemical, and pharmacological data.
The goal of the database is to make these data accessible to researchers worldwide to help with hypothesis generation and support drug discovery decisions.
Users can search canSAR using text queries, protein/gene name searches, any keyword searches, chemical structure searches, and sequence similarity searches. Users can also explore and filter chemical compound sets, view experimental data, and produce summary plots.
The canSAR database was launched in 2011 with the goal of using Big Data approaches to build a detailed picture of how the majority of known human molecules behave.
The database has already collated billions of experimental measurements, mapping the actions of 1 million drugs and chemicals on human proteins, and it has combined these data with genetic information and results from clinical trials.
The updated version of canSAR uses artificial intelligence to identify nooks and crannies on the surface of faulty cancer-causing molecules as a key step in designing new drugs to block them. It also allows researchers to identify communication lines that can be intercepted within tumor cells, opening up potential new approaches for cancer treatment.
The growing database now holds the 3D structures of almost 3 million cavities on the surface of nearly 110,000 molecules.
“Our database is constantly growing with information and is the largest of its kind, with more than 140,000 users from over 175 countries,” said Bissan Al-Lazikani, PhD, of The Institute of Cancer Research in London, UK.
“And we regularly develop new artificial intelligence technologies that help scientists make predictions and design experiments. Our aim is that cancer scientists will be armed with the data they need to carry out life-saving research into the most exciting drugs of the future.”
“Scientists need to find all the information there is about a faulty gene or protein to understand whether a new drug might work. These data are vast and scattered, but the canSAR database brings them together and adds value by identifying hidden links and presenting the key information easily.”
Details on the updates to canSAR have been published in Nucleic Acid Research. The database is available online at https://cansar.icr.ac.uk/.

Photo by Rhoda Baer
Researchers have updated the canSAR database, a tool designed to aid cancer drug discovery, by adding 3D structures of faulty proteins and maps of cancer’s communication networks.
The canSAR database brings together biological, chemical, and pharmacological data.
The goal of the database is to make these data accessible to researchers worldwide to help with hypothesis generation and support drug discovery decisions.
Users can search canSAR using text queries, protein/gene name searches, any keyword searches, chemical structure searches, and sequence similarity searches. Users can also explore and filter chemical compound sets, view experimental data, and produce summary plots.
The canSAR database was launched in 2011 with the goal of using Big Data approaches to build a detailed picture of how the majority of known human molecules behave.
The database has already collated billions of experimental measurements, mapping the actions of 1 million drugs and chemicals on human proteins, and it has combined these data with genetic information and results from clinical trials.
The updated version of canSAR uses artificial intelligence to identify nooks and crannies on the surface of faulty cancer-causing molecules as a key step in designing new drugs to block them. It also allows researchers to identify communication lines that can be intercepted within tumor cells, opening up potential new approaches for cancer treatment.
The growing database now holds the 3D structures of almost 3 million cavities on the surface of nearly 110,000 molecules.
“Our database is constantly growing with information and is the largest of its kind, with more than 140,000 users from over 175 countries,” said Bissan Al-Lazikani, PhD, of The Institute of Cancer Research in London, UK.
“And we regularly develop new artificial intelligence technologies that help scientists make predictions and design experiments. Our aim is that cancer scientists will be armed with the data they need to carry out life-saving research into the most exciting drugs of the future.”
“Scientists need to find all the information there is about a faulty gene or protein to understand whether a new drug might work. These data are vast and scattered, but the canSAR database brings them together and adds value by identifying hidden links and presenting the key information easily.”
Details on the updates to canSAR have been published in Nucleic Acid Research. The database is available online at https://cansar.icr.ac.uk/.

Photo by Rhoda Baer
Researchers have updated the canSAR database, a tool designed to aid cancer drug discovery, by adding 3D structures of faulty proteins and maps of cancer’s communication networks.
The canSAR database brings together biological, chemical, and pharmacological data.
The goal of the database is to make these data accessible to researchers worldwide to help with hypothesis generation and support drug discovery decisions.
Users can search canSAR using text queries, protein/gene name searches, any keyword searches, chemical structure searches, and sequence similarity searches. Users can also explore and filter chemical compound sets, view experimental data, and produce summary plots.
The canSAR database was launched in 2011 with the goal of using Big Data approaches to build a detailed picture of how the majority of known human molecules behave.
The database has already collated billions of experimental measurements, mapping the actions of 1 million drugs and chemicals on human proteins, and it has combined these data with genetic information and results from clinical trials.
The updated version of canSAR uses artificial intelligence to identify nooks and crannies on the surface of faulty cancer-causing molecules as a key step in designing new drugs to block them. It also allows researchers to identify communication lines that can be intercepted within tumor cells, opening up potential new approaches for cancer treatment.
The growing database now holds the 3D structures of almost 3 million cavities on the surface of nearly 110,000 molecules.
“Our database is constantly growing with information and is the largest of its kind, with more than 140,000 users from over 175 countries,” said Bissan Al-Lazikani, PhD, of The Institute of Cancer Research in London, UK.
“And we regularly develop new artificial intelligence technologies that help scientists make predictions and design experiments. Our aim is that cancer scientists will be armed with the data they need to carry out life-saving research into the most exciting drugs of the future.”
“Scientists need to find all the information there is about a faulty gene or protein to understand whether a new drug might work. These data are vast and scattered, but the canSAR database brings them together and adds value by identifying hidden links and presenting the key information easily.”
Details on the updates to canSAR have been published in Nucleic Acid Research. The database is available online at https://cansar.icr.ac.uk/.