User login
CCJM delivers practical clinical articles relevant to internists, cardiologists, endocrinologists, and other specialists, all written by known experts.
Copyright © 2019 Cleveland Clinic. All rights reserved. The information provided is for educational purposes only. Use of this website is subject to the disclaimer and privacy policy.
gambling
compulsive behaviors
ammunition
assault rifle
black jack
Boko Haram
bondage
child abuse
cocaine
Daech
drug paraphernalia
explosion
gun
human trafficking
ISIL
ISIS
Islamic caliphate
Islamic state
mixed martial arts
MMA
molestation
national rifle association
NRA
nsfw
pedophile
pedophilia
poker
porn
pornography
psychedelic drug
recreational drug
sex slave rings
slot machine
terrorism
terrorist
Texas hold 'em
UFC
substance abuse
abuseed
abuseer
abusees
abuseing
abusely
abuses
aeolus
aeolused
aeoluser
aeoluses
aeolusing
aeolusly
aeoluss
ahole
aholeed
aholeer
aholees
aholeing
aholely
aholes
alcohol
alcoholed
alcoholer
alcoholes
alcoholing
alcoholly
alcohols
allman
allmaned
allmaner
allmanes
allmaning
allmanly
allmans
alted
altes
alting
altly
alts
analed
analer
anales
analing
anally
analprobe
analprobeed
analprobeer
analprobees
analprobeing
analprobely
analprobes
anals
anilingus
anilingused
anilinguser
anilinguses
anilingusing
anilingusly
anilinguss
anus
anused
anuser
anuses
anusing
anusly
anuss
areola
areolaed
areolaer
areolaes
areolaing
areolaly
areolas
areole
areoleed
areoleer
areolees
areoleing
areolely
areoles
arian
arianed
arianer
arianes
arianing
arianly
arians
aryan
aryaned
aryaner
aryanes
aryaning
aryanly
aryans
asiaed
asiaer
asiaes
asiaing
asialy
asias
ass
ass hole
ass lick
ass licked
ass licker
ass lickes
ass licking
ass lickly
ass licks
assbang
assbanged
assbangeded
assbangeder
assbangedes
assbangeding
assbangedly
assbangeds
assbanger
assbanges
assbanging
assbangly
assbangs
assbangsed
assbangser
assbangses
assbangsing
assbangsly
assbangss
assed
asser
asses
assesed
asseser
asseses
assesing
assesly
assess
assfuck
assfucked
assfucker
assfuckered
assfuckerer
assfuckeres
assfuckering
assfuckerly
assfuckers
assfuckes
assfucking
assfuckly
assfucks
asshat
asshated
asshater
asshates
asshating
asshatly
asshats
assholeed
assholeer
assholees
assholeing
assholely
assholes
assholesed
assholeser
assholeses
assholesing
assholesly
assholess
assing
assly
assmaster
assmastered
assmasterer
assmasteres
assmastering
assmasterly
assmasters
assmunch
assmunched
assmuncher
assmunches
assmunching
assmunchly
assmunchs
asss
asswipe
asswipeed
asswipeer
asswipees
asswipeing
asswipely
asswipes
asswipesed
asswipeser
asswipeses
asswipesing
asswipesly
asswipess
azz
azzed
azzer
azzes
azzing
azzly
azzs
babeed
babeer
babees
babeing
babely
babes
babesed
babeser
babeses
babesing
babesly
babess
ballsac
ballsaced
ballsacer
ballsaces
ballsacing
ballsack
ballsacked
ballsacker
ballsackes
ballsacking
ballsackly
ballsacks
ballsacly
ballsacs
ballsed
ballser
ballses
ballsing
ballsly
ballss
barf
barfed
barfer
barfes
barfing
barfly
barfs
bastard
bastarded
bastarder
bastardes
bastarding
bastardly
bastards
bastardsed
bastardser
bastardses
bastardsing
bastardsly
bastardss
bawdy
bawdyed
bawdyer
bawdyes
bawdying
bawdyly
bawdys
beaner
beanered
beanerer
beaneres
beanering
beanerly
beaners
beardedclam
beardedclamed
beardedclamer
beardedclames
beardedclaming
beardedclamly
beardedclams
beastiality
beastialityed
beastialityer
beastialityes
beastialitying
beastialityly
beastialitys
beatch
beatched
beatcher
beatches
beatching
beatchly
beatchs
beater
beatered
beaterer
beateres
beatering
beaterly
beaters
beered
beerer
beeres
beering
beerly
beeyotch
beeyotched
beeyotcher
beeyotches
beeyotching
beeyotchly
beeyotchs
beotch
beotched
beotcher
beotches
beotching
beotchly
beotchs
biatch
biatched
biatcher
biatches
biatching
biatchly
biatchs
big tits
big titsed
big titser
big titses
big titsing
big titsly
big titss
bigtits
bigtitsed
bigtitser
bigtitses
bigtitsing
bigtitsly
bigtitss
bimbo
bimboed
bimboer
bimboes
bimboing
bimboly
bimbos
bisexualed
bisexualer
bisexuales
bisexualing
bisexually
bisexuals
bitch
bitched
bitcheded
bitcheder
bitchedes
bitcheding
bitchedly
bitcheds
bitcher
bitches
bitchesed
bitcheser
bitcheses
bitchesing
bitchesly
bitchess
bitching
bitchly
bitchs
bitchy
bitchyed
bitchyer
bitchyes
bitchying
bitchyly
bitchys
bleached
bleacher
bleaches
bleaching
bleachly
bleachs
blow job
blow jobed
blow jober
blow jobes
blow jobing
blow jobly
blow jobs
blowed
blower
blowes
blowing
blowjob
blowjobed
blowjober
blowjobes
blowjobing
blowjobly
blowjobs
blowjobsed
blowjobser
blowjobses
blowjobsing
blowjobsly
blowjobss
blowly
blows
boink
boinked
boinker
boinkes
boinking
boinkly
boinks
bollock
bollocked
bollocker
bollockes
bollocking
bollockly
bollocks
bollocksed
bollockser
bollockses
bollocksing
bollocksly
bollockss
bollok
bolloked
bolloker
bollokes
bolloking
bollokly
bolloks
boner
bonered
bonerer
boneres
bonering
bonerly
boners
bonersed
bonerser
bonerses
bonersing
bonersly
bonerss
bong
bonged
bonger
bonges
bonging
bongly
bongs
boob
boobed
boober
boobes
boobies
boobiesed
boobieser
boobieses
boobiesing
boobiesly
boobiess
boobing
boobly
boobs
boobsed
boobser
boobses
boobsing
boobsly
boobss
booby
boobyed
boobyer
boobyes
boobying
boobyly
boobys
booger
boogered
boogerer
boogeres
boogering
boogerly
boogers
bookie
bookieed
bookieer
bookiees
bookieing
bookiely
bookies
bootee
booteeed
booteeer
booteees
booteeing
booteely
bootees
bootie
bootieed
bootieer
bootiees
bootieing
bootiely
booties
booty
bootyed
bootyer
bootyes
bootying
bootyly
bootys
boozeed
boozeer
boozees
boozeing
boozely
boozer
boozered
boozerer
boozeres
boozering
boozerly
boozers
boozes
boozy
boozyed
boozyer
boozyes
boozying
boozyly
boozys
bosomed
bosomer
bosomes
bosoming
bosomly
bosoms
bosomy
bosomyed
bosomyer
bosomyes
bosomying
bosomyly
bosomys
bugger
buggered
buggerer
buggeres
buggering
buggerly
buggers
bukkake
bukkakeed
bukkakeer
bukkakees
bukkakeing
bukkakely
bukkakes
bull shit
bull shited
bull shiter
bull shites
bull shiting
bull shitly
bull shits
bullshit
bullshited
bullshiter
bullshites
bullshiting
bullshitly
bullshits
bullshitsed
bullshitser
bullshitses
bullshitsing
bullshitsly
bullshitss
bullshitted
bullshitteded
bullshitteder
bullshittedes
bullshitteding
bullshittedly
bullshitteds
bullturds
bullturdsed
bullturdser
bullturdses
bullturdsing
bullturdsly
bullturdss
bung
bunged
bunger
bunges
bunging
bungly
bungs
busty
bustyed
bustyer
bustyes
bustying
bustyly
bustys
butt
butt fuck
butt fucked
butt fucker
butt fuckes
butt fucking
butt fuckly
butt fucks
butted
buttes
buttfuck
buttfucked
buttfucker
buttfuckered
buttfuckerer
buttfuckeres
buttfuckering
buttfuckerly
buttfuckers
buttfuckes
buttfucking
buttfuckly
buttfucks
butting
buttly
buttplug
buttpluged
buttpluger
buttpluges
buttpluging
buttplugly
buttplugs
butts
caca
cacaed
cacaer
cacaes
cacaing
cacaly
cacas
cahone
cahoneed
cahoneer
cahonees
cahoneing
cahonely
cahones
cameltoe
cameltoeed
cameltoeer
cameltoees
cameltoeing
cameltoely
cameltoes
carpetmuncher
carpetmunchered
carpetmuncherer
carpetmuncheres
carpetmunchering
carpetmuncherly
carpetmunchers
cawk
cawked
cawker
cawkes
cawking
cawkly
cawks
chinc
chinced
chincer
chinces
chincing
chincly
chincs
chincsed
chincser
chincses
chincsing
chincsly
chincss
chink
chinked
chinker
chinkes
chinking
chinkly
chinks
chode
chodeed
chodeer
chodees
chodeing
chodely
chodes
chodesed
chodeser
chodeses
chodesing
chodesly
chodess
clit
clited
cliter
clites
cliting
clitly
clitoris
clitorised
clitoriser
clitorises
clitorising
clitorisly
clitoriss
clitorus
clitorused
clitoruser
clitoruses
clitorusing
clitorusly
clitoruss
clits
clitsed
clitser
clitses
clitsing
clitsly
clitss
clitty
clittyed
clittyer
clittyes
clittying
clittyly
clittys
cocain
cocaine
cocained
cocaineed
cocaineer
cocainees
cocaineing
cocainely
cocainer
cocaines
cocaining
cocainly
cocains
cock
cock sucker
cock suckered
cock suckerer
cock suckeres
cock suckering
cock suckerly
cock suckers
cockblock
cockblocked
cockblocker
cockblockes
cockblocking
cockblockly
cockblocks
cocked
cocker
cockes
cockholster
cockholstered
cockholsterer
cockholsteres
cockholstering
cockholsterly
cockholsters
cocking
cockknocker
cockknockered
cockknockerer
cockknockeres
cockknockering
cockknockerly
cockknockers
cockly
cocks
cocksed
cockser
cockses
cocksing
cocksly
cocksmoker
cocksmokered
cocksmokerer
cocksmokeres
cocksmokering
cocksmokerly
cocksmokers
cockss
cocksucker
cocksuckered
cocksuckerer
cocksuckeres
cocksuckering
cocksuckerly
cocksuckers
coital
coitaled
coitaler
coitales
coitaling
coitally
coitals
commie
commieed
commieer
commiees
commieing
commiely
commies
condomed
condomer
condomes
condoming
condomly
condoms
coon
cooned
cooner
coones
cooning
coonly
coons
coonsed
coonser
coonses
coonsing
coonsly
coonss
corksucker
corksuckered
corksuckerer
corksuckeres
corksuckering
corksuckerly
corksuckers
cracked
crackwhore
crackwhoreed
crackwhoreer
crackwhorees
crackwhoreing
crackwhorely
crackwhores
crap
craped
craper
crapes
craping
craply
crappy
crappyed
crappyer
crappyes
crappying
crappyly
crappys
cum
cumed
cumer
cumes
cuming
cumly
cummin
cummined
cumminer
cummines
cumming
cumminged
cumminger
cumminges
cumminging
cummingly
cummings
cummining
cumminly
cummins
cums
cumshot
cumshoted
cumshoter
cumshotes
cumshoting
cumshotly
cumshots
cumshotsed
cumshotser
cumshotses
cumshotsing
cumshotsly
cumshotss
cumslut
cumsluted
cumsluter
cumslutes
cumsluting
cumslutly
cumsluts
cumstain
cumstained
cumstainer
cumstaines
cumstaining
cumstainly
cumstains
cunilingus
cunilingused
cunilinguser
cunilinguses
cunilingusing
cunilingusly
cunilinguss
cunnilingus
cunnilingused
cunnilinguser
cunnilinguses
cunnilingusing
cunnilingusly
cunnilinguss
cunny
cunnyed
cunnyer
cunnyes
cunnying
cunnyly
cunnys
cunt
cunted
cunter
cuntes
cuntface
cuntfaceed
cuntfaceer
cuntfacees
cuntfaceing
cuntfacely
cuntfaces
cunthunter
cunthuntered
cunthunterer
cunthunteres
cunthuntering
cunthunterly
cunthunters
cunting
cuntlick
cuntlicked
cuntlicker
cuntlickered
cuntlickerer
cuntlickeres
cuntlickering
cuntlickerly
cuntlickers
cuntlickes
cuntlicking
cuntlickly
cuntlicks
cuntly
cunts
cuntsed
cuntser
cuntses
cuntsing
cuntsly
cuntss
dago
dagoed
dagoer
dagoes
dagoing
dagoly
dagos
dagosed
dagoser
dagoses
dagosing
dagosly
dagoss
dammit
dammited
dammiter
dammites
dammiting
dammitly
dammits
damn
damned
damneded
damneder
damnedes
damneding
damnedly
damneds
damner
damnes
damning
damnit
damnited
damniter
damnites
damniting
damnitly
damnits
damnly
damns
dick
dickbag
dickbaged
dickbager
dickbages
dickbaging
dickbagly
dickbags
dickdipper
dickdippered
dickdipperer
dickdipperes
dickdippering
dickdipperly
dickdippers
dicked
dicker
dickes
dickface
dickfaceed
dickfaceer
dickfacees
dickfaceing
dickfacely
dickfaces
dickflipper
dickflippered
dickflipperer
dickflipperes
dickflippering
dickflipperly
dickflippers
dickhead
dickheaded
dickheader
dickheades
dickheading
dickheadly
dickheads
dickheadsed
dickheadser
dickheadses
dickheadsing
dickheadsly
dickheadss
dicking
dickish
dickished
dickisher
dickishes
dickishing
dickishly
dickishs
dickly
dickripper
dickrippered
dickripperer
dickripperes
dickrippering
dickripperly
dickrippers
dicks
dicksipper
dicksippered
dicksipperer
dicksipperes
dicksippering
dicksipperly
dicksippers
dickweed
dickweeded
dickweeder
dickweedes
dickweeding
dickweedly
dickweeds
dickwhipper
dickwhippered
dickwhipperer
dickwhipperes
dickwhippering
dickwhipperly
dickwhippers
dickzipper
dickzippered
dickzipperer
dickzipperes
dickzippering
dickzipperly
dickzippers
diddle
diddleed
diddleer
diddlees
diddleing
diddlely
diddles
dike
dikeed
dikeer
dikees
dikeing
dikely
dikes
dildo
dildoed
dildoer
dildoes
dildoing
dildoly
dildos
dildosed
dildoser
dildoses
dildosing
dildosly
dildoss
diligaf
diligafed
diligafer
diligafes
diligafing
diligafly
diligafs
dillweed
dillweeded
dillweeder
dillweedes
dillweeding
dillweedly
dillweeds
dimwit
dimwited
dimwiter
dimwites
dimwiting
dimwitly
dimwits
dingle
dingleed
dingleer
dinglees
dingleing
dinglely
dingles
dipship
dipshiped
dipshiper
dipshipes
dipshiping
dipshiply
dipships
dizzyed
dizzyer
dizzyes
dizzying
dizzyly
dizzys
doggiestyleed
doggiestyleer
doggiestylees
doggiestyleing
doggiestylely
doggiestyles
doggystyleed
doggystyleer
doggystylees
doggystyleing
doggystylely
doggystyles
dong
donged
donger
donges
donging
dongly
dongs
doofus
doofused
doofuser
doofuses
doofusing
doofusly
doofuss
doosh
dooshed
doosher
dooshes
dooshing
dooshly
dooshs
dopeyed
dopeyer
dopeyes
dopeying
dopeyly
dopeys
douchebag
douchebaged
douchebager
douchebages
douchebaging
douchebagly
douchebags
douchebagsed
douchebagser
douchebagses
douchebagsing
douchebagsly
douchebagss
doucheed
doucheer
douchees
doucheing
douchely
douches
douchey
doucheyed
doucheyer
doucheyes
doucheying
doucheyly
doucheys
drunk
drunked
drunker
drunkes
drunking
drunkly
drunks
dumass
dumassed
dumasser
dumasses
dumassing
dumassly
dumasss
dumbass
dumbassed
dumbasser
dumbasses
dumbassesed
dumbasseser
dumbasseses
dumbassesing
dumbassesly
dumbassess
dumbassing
dumbassly
dumbasss
dummy
dummyed
dummyer
dummyes
dummying
dummyly
dummys
dyke
dykeed
dykeer
dykees
dykeing
dykely
dykes
dykesed
dykeser
dykeses
dykesing
dykesly
dykess
erotic
eroticed
eroticer
erotices
eroticing
eroticly
erotics
extacy
extacyed
extacyer
extacyes
extacying
extacyly
extacys
extasy
extasyed
extasyer
extasyes
extasying
extasyly
extasys
fack
facked
facker
fackes
facking
fackly
facks
fag
faged
fager
fages
fagg
fagged
faggeded
faggeder
faggedes
faggeding
faggedly
faggeds
fagger
fagges
fagging
faggit
faggited
faggiter
faggites
faggiting
faggitly
faggits
faggly
faggot
faggoted
faggoter
faggotes
faggoting
faggotly
faggots
faggs
faging
fagly
fagot
fagoted
fagoter
fagotes
fagoting
fagotly
fagots
fags
fagsed
fagser
fagses
fagsing
fagsly
fagss
faig
faiged
faiger
faiges
faiging
faigly
faigs
faigt
faigted
faigter
faigtes
faigting
faigtly
faigts
fannybandit
fannybandited
fannybanditer
fannybandites
fannybanditing
fannybanditly
fannybandits
farted
farter
fartes
farting
fartknocker
fartknockered
fartknockerer
fartknockeres
fartknockering
fartknockerly
fartknockers
fartly
farts
felch
felched
felcher
felchered
felcherer
felcheres
felchering
felcherly
felchers
felches
felching
felchinged
felchinger
felchinges
felchinging
felchingly
felchings
felchly
felchs
fellate
fellateed
fellateer
fellatees
fellateing
fellately
fellates
fellatio
fellatioed
fellatioer
fellatioes
fellatioing
fellatioly
fellatios
feltch
feltched
feltcher
feltchered
feltcherer
feltcheres
feltchering
feltcherly
feltchers
feltches
feltching
feltchly
feltchs
feom
feomed
feomer
feomes
feoming
feomly
feoms
fisted
fisteded
fisteder
fistedes
fisteding
fistedly
fisteds
fisting
fistinged
fistinger
fistinges
fistinging
fistingly
fistings
fisty
fistyed
fistyer
fistyes
fistying
fistyly
fistys
floozy
floozyed
floozyer
floozyes
floozying
floozyly
floozys
foad
foaded
foader
foades
foading
foadly
foads
fondleed
fondleer
fondlees
fondleing
fondlely
fondles
foobar
foobared
foobarer
foobares
foobaring
foobarly
foobars
freex
freexed
freexer
freexes
freexing
freexly
freexs
frigg
frigga
friggaed
friggaer
friggaes
friggaing
friggaly
friggas
frigged
frigger
frigges
frigging
friggly
friggs
fubar
fubared
fubarer
fubares
fubaring
fubarly
fubars
fuck
fuckass
fuckassed
fuckasser
fuckasses
fuckassing
fuckassly
fuckasss
fucked
fuckeded
fuckeder
fuckedes
fuckeding
fuckedly
fuckeds
fucker
fuckered
fuckerer
fuckeres
fuckering
fuckerly
fuckers
fuckes
fuckface
fuckfaceed
fuckfaceer
fuckfacees
fuckfaceing
fuckfacely
fuckfaces
fuckin
fuckined
fuckiner
fuckines
fucking
fuckinged
fuckinger
fuckinges
fuckinging
fuckingly
fuckings
fuckining
fuckinly
fuckins
fuckly
fucknugget
fucknuggeted
fucknuggeter
fucknuggetes
fucknuggeting
fucknuggetly
fucknuggets
fucknut
fucknuted
fucknuter
fucknutes
fucknuting
fucknutly
fucknuts
fuckoff
fuckoffed
fuckoffer
fuckoffes
fuckoffing
fuckoffly
fuckoffs
fucks
fucksed
fuckser
fuckses
fucksing
fucksly
fuckss
fucktard
fucktarded
fucktarder
fucktardes
fucktarding
fucktardly
fucktards
fuckup
fuckuped
fuckuper
fuckupes
fuckuping
fuckuply
fuckups
fuckwad
fuckwaded
fuckwader
fuckwades
fuckwading
fuckwadly
fuckwads
fuckwit
fuckwited
fuckwiter
fuckwites
fuckwiting
fuckwitly
fuckwits
fudgepacker
fudgepackered
fudgepackerer
fudgepackeres
fudgepackering
fudgepackerly
fudgepackers
fuk
fuked
fuker
fukes
fuking
fukly
fuks
fvck
fvcked
fvcker
fvckes
fvcking
fvckly
fvcks
fxck
fxcked
fxcker
fxckes
fxcking
fxckly
fxcks
gae
gaeed
gaeer
gaees
gaeing
gaely
gaes
gai
gaied
gaier
gaies
gaiing
gaily
gais
ganja
ganjaed
ganjaer
ganjaes
ganjaing
ganjaly
ganjas
gayed
gayer
gayes
gaying
gayly
gays
gaysed
gayser
gayses
gaysing
gaysly
gayss
gey
geyed
geyer
geyes
geying
geyly
geys
gfc
gfced
gfcer
gfces
gfcing
gfcly
gfcs
gfy
gfyed
gfyer
gfyes
gfying
gfyly
gfys
ghay
ghayed
ghayer
ghayes
ghaying
ghayly
ghays
ghey
gheyed
gheyer
gheyes
gheying
gheyly
gheys
gigolo
gigoloed
gigoloer
gigoloes
gigoloing
gigololy
gigolos
goatse
goatseed
goatseer
goatsees
goatseing
goatsely
goatses
godamn
godamned
godamner
godamnes
godamning
godamnit
godamnited
godamniter
godamnites
godamniting
godamnitly
godamnits
godamnly
godamns
goddam
goddamed
goddamer
goddames
goddaming
goddamly
goddammit
goddammited
goddammiter
goddammites
goddammiting
goddammitly
goddammits
goddamn
goddamned
goddamner
goddamnes
goddamning
goddamnly
goddamns
goddams
goldenshower
goldenshowered
goldenshowerer
goldenshoweres
goldenshowering
goldenshowerly
goldenshowers
gonad
gonaded
gonader
gonades
gonading
gonadly
gonads
gonadsed
gonadser
gonadses
gonadsing
gonadsly
gonadss
gook
gooked
gooker
gookes
gooking
gookly
gooks
gooksed
gookser
gookses
gooksing
gooksly
gookss
gringo
gringoed
gringoer
gringoes
gringoing
gringoly
gringos
gspot
gspoted
gspoter
gspotes
gspoting
gspotly
gspots
gtfo
gtfoed
gtfoer
gtfoes
gtfoing
gtfoly
gtfos
guido
guidoed
guidoer
guidoes
guidoing
guidoly
guidos
handjob
handjobed
handjober
handjobes
handjobing
handjobly
handjobs
hard on
hard oned
hard oner
hard ones
hard oning
hard only
hard ons
hardknight
hardknighted
hardknighter
hardknightes
hardknighting
hardknightly
hardknights
hebe
hebeed
hebeer
hebees
hebeing
hebely
hebes
heeb
heebed
heeber
heebes
heebing
heebly
heebs
hell
helled
heller
helles
helling
hellly
hells
hemp
hemped
hemper
hempes
hemping
hemply
hemps
heroined
heroiner
heroines
heroining
heroinly
heroins
herp
herped
herper
herpes
herpesed
herpeser
herpeses
herpesing
herpesly
herpess
herping
herply
herps
herpy
herpyed
herpyer
herpyes
herpying
herpyly
herpys
hitler
hitlered
hitlerer
hitleres
hitlering
hitlerly
hitlers
hived
hiver
hives
hiving
hivly
hivs
hobag
hobaged
hobager
hobages
hobaging
hobagly
hobags
homey
homeyed
homeyer
homeyes
homeying
homeyly
homeys
homo
homoed
homoer
homoes
homoey
homoeyed
homoeyer
homoeyes
homoeying
homoeyly
homoeys
homoing
homoly
homos
honky
honkyed
honkyer
honkyes
honkying
honkyly
honkys
hooch
hooched
hoocher
hooches
hooching
hoochly
hoochs
hookah
hookahed
hookaher
hookahes
hookahing
hookahly
hookahs
hooker
hookered
hookerer
hookeres
hookering
hookerly
hookers
hoor
hoored
hoorer
hoores
hooring
hoorly
hoors
hootch
hootched
hootcher
hootches
hootching
hootchly
hootchs
hooter
hootered
hooterer
hooteres
hootering
hooterly
hooters
hootersed
hooterser
hooterses
hootersing
hootersly
hooterss
horny
hornyed
hornyer
hornyes
hornying
hornyly
hornys
houstoned
houstoner
houstones
houstoning
houstonly
houstons
hump
humped
humpeded
humpeder
humpedes
humpeding
humpedly
humpeds
humper
humpes
humping
humpinged
humpinger
humpinges
humpinging
humpingly
humpings
humply
humps
husbanded
husbander
husbandes
husbanding
husbandly
husbands
hussy
hussyed
hussyer
hussyes
hussying
hussyly
hussys
hymened
hymener
hymenes
hymening
hymenly
hymens
inbred
inbreded
inbreder
inbredes
inbreding
inbredly
inbreds
incest
incested
incester
incestes
incesting
incestly
incests
injun
injuned
injuner
injunes
injuning
injunly
injuns
jackass
jackassed
jackasser
jackasses
jackassing
jackassly
jackasss
jackhole
jackholeed
jackholeer
jackholees
jackholeing
jackholely
jackholes
jackoff
jackoffed
jackoffer
jackoffes
jackoffing
jackoffly
jackoffs
jap
japed
japer
japes
japing
japly
japs
japsed
japser
japses
japsing
japsly
japss
jerkoff
jerkoffed
jerkoffer
jerkoffes
jerkoffing
jerkoffly
jerkoffs
jerks
jism
jismed
jismer
jismes
jisming
jismly
jisms
jiz
jized
jizer
jizes
jizing
jizly
jizm
jizmed
jizmer
jizmes
jizming
jizmly
jizms
jizs
jizz
jizzed
jizzeded
jizzeder
jizzedes
jizzeding
jizzedly
jizzeds
jizzer
jizzes
jizzing
jizzly
jizzs
junkie
junkieed
junkieer
junkiees
junkieing
junkiely
junkies
junky
junkyed
junkyer
junkyes
junkying
junkyly
junkys
kike
kikeed
kikeer
kikees
kikeing
kikely
kikes
kikesed
kikeser
kikeses
kikesing
kikesly
kikess
killed
killer
killes
killing
killly
kills
kinky
kinkyed
kinkyer
kinkyes
kinkying
kinkyly
kinkys
kkk
kkked
kkker
kkkes
kkking
kkkly
kkks
klan
klaned
klaner
klanes
klaning
klanly
klans
knobend
knobended
knobender
knobendes
knobending
knobendly
knobends
kooch
kooched
koocher
kooches
koochesed
koocheser
koocheses
koochesing
koochesly
koochess
kooching
koochly
koochs
kootch
kootched
kootcher
kootches
kootching
kootchly
kootchs
kraut
krauted
krauter
krautes
krauting
krautly
krauts
kyke
kykeed
kykeer
kykees
kykeing
kykely
kykes
lech
leched
lecher
leches
leching
lechly
lechs
leper
lepered
leperer
leperes
lepering
leperly
lepers
lesbiansed
lesbianser
lesbianses
lesbiansing
lesbiansly
lesbianss
lesbo
lesboed
lesboer
lesboes
lesboing
lesboly
lesbos
lesbosed
lesboser
lesboses
lesbosing
lesbosly
lesboss
lez
lezbianed
lezbianer
lezbianes
lezbianing
lezbianly
lezbians
lezbiansed
lezbianser
lezbianses
lezbiansing
lezbiansly
lezbianss
lezbo
lezboed
lezboer
lezboes
lezboing
lezboly
lezbos
lezbosed
lezboser
lezboses
lezbosing
lezbosly
lezboss
lezed
lezer
lezes
lezing
lezly
lezs
lezzie
lezzieed
lezzieer
lezziees
lezzieing
lezziely
lezzies
lezziesed
lezzieser
lezzieses
lezziesing
lezziesly
lezziess
lezzy
lezzyed
lezzyer
lezzyes
lezzying
lezzyly
lezzys
lmaoed
lmaoer
lmaoes
lmaoing
lmaoly
lmaos
lmfao
lmfaoed
lmfaoer
lmfaoes
lmfaoing
lmfaoly
lmfaos
loined
loiner
loines
loining
loinly
loins
loinsed
loinser
loinses
loinsing
loinsly
loinss
lubeed
lubeer
lubees
lubeing
lubely
lubes
lusty
lustyed
lustyer
lustyes
lustying
lustyly
lustys
massa
massaed
massaer
massaes
massaing
massaly
massas
masterbate
masterbateed
masterbateer
masterbatees
masterbateing
masterbately
masterbates
masterbating
masterbatinged
masterbatinger
masterbatinges
masterbatinging
masterbatingly
masterbatings
masterbation
masterbationed
masterbationer
masterbationes
masterbationing
masterbationly
masterbations
masturbate
masturbateed
masturbateer
masturbatees
masturbateing
masturbately
masturbates
masturbating
masturbatinged
masturbatinger
masturbatinges
masturbatinging
masturbatingly
masturbatings
masturbation
masturbationed
masturbationer
masturbationes
masturbationing
masturbationly
masturbations
methed
mether
methes
mething
methly
meths
militaryed
militaryer
militaryes
militarying
militaryly
militarys
mofo
mofoed
mofoer
mofoes
mofoing
mofoly
mofos
molest
molested
molester
molestes
molesting
molestly
molests
moolie
moolieed
moolieer
mooliees
moolieing
mooliely
moolies
moron
moroned
moroner
morones
moroning
moronly
morons
motherfucka
motherfuckaed
motherfuckaer
motherfuckaes
motherfuckaing
motherfuckaly
motherfuckas
motherfucker
motherfuckered
motherfuckerer
motherfuckeres
motherfuckering
motherfuckerly
motherfuckers
motherfucking
motherfuckinged
motherfuckinger
motherfuckinges
motherfuckinging
motherfuckingly
motherfuckings
mtherfucker
mtherfuckered
mtherfuckerer
mtherfuckeres
mtherfuckering
mtherfuckerly
mtherfuckers
mthrfucker
mthrfuckered
mthrfuckerer
mthrfuckeres
mthrfuckering
mthrfuckerly
mthrfuckers
mthrfucking
mthrfuckinged
mthrfuckinger
mthrfuckinges
mthrfuckinging
mthrfuckingly
mthrfuckings
muff
muffdiver
muffdivered
muffdiverer
muffdiveres
muffdivering
muffdiverly
muffdivers
muffed
muffer
muffes
muffing
muffly
muffs
murdered
murderer
murderes
murdering
murderly
murders
muthafuckaz
muthafuckazed
muthafuckazer
muthafuckazes
muthafuckazing
muthafuckazly
muthafuckazs
muthafucker
muthafuckered
muthafuckerer
muthafuckeres
muthafuckering
muthafuckerly
muthafuckers
mutherfucker
mutherfuckered
mutherfuckerer
mutherfuckeres
mutherfuckering
mutherfuckerly
mutherfuckers
mutherfucking
mutherfuckinged
mutherfuckinger
mutherfuckinges
mutherfuckinging
mutherfuckingly
mutherfuckings
muthrfucking
muthrfuckinged
muthrfuckinger
muthrfuckinges
muthrfuckinging
muthrfuckingly
muthrfuckings
nad
naded
nader
nades
nading
nadly
nads
nadsed
nadser
nadses
nadsing
nadsly
nadss
nakeded
nakeder
nakedes
nakeding
nakedly
nakeds
napalm
napalmed
napalmer
napalmes
napalming
napalmly
napalms
nappy
nappyed
nappyer
nappyes
nappying
nappyly
nappys
nazi
nazied
nazier
nazies
naziing
nazily
nazis
nazism
nazismed
nazismer
nazismes
nazisming
nazismly
nazisms
negro
negroed
negroer
negroes
negroing
negroly
negros
nigga
niggaed
niggaer
niggaes
niggah
niggahed
niggaher
niggahes
niggahing
niggahly
niggahs
niggaing
niggaly
niggas
niggased
niggaser
niggases
niggasing
niggasly
niggass
niggaz
niggazed
niggazer
niggazes
niggazing
niggazly
niggazs
nigger
niggered
niggerer
niggeres
niggering
niggerly
niggers
niggersed
niggerser
niggerses
niggersing
niggersly
niggerss
niggle
niggleed
niggleer
nigglees
niggleing
nigglely
niggles
niglet
nigleted
nigleter
nigletes
nigleting
nigletly
niglets
nimrod
nimroded
nimroder
nimrodes
nimroding
nimrodly
nimrods
ninny
ninnyed
ninnyer
ninnyes
ninnying
ninnyly
ninnys
nooky
nookyed
nookyer
nookyes
nookying
nookyly
nookys
nuccitelli
nuccitellied
nuccitellier
nuccitellies
nuccitelliing
nuccitellily
nuccitellis
nympho
nymphoed
nymphoer
nymphoes
nymphoing
nympholy
nymphos
opium
opiumed
opiumer
opiumes
opiuming
opiumly
opiums
orgies
orgiesed
orgieser
orgieses
orgiesing
orgiesly
orgiess
orgy
orgyed
orgyer
orgyes
orgying
orgyly
orgys
paddy
paddyed
paddyer
paddyes
paddying
paddyly
paddys
paki
pakied
pakier
pakies
pakiing
pakily
pakis
pantie
pantieed
pantieer
pantiees
pantieing
pantiely
panties
pantiesed
pantieser
pantieses
pantiesing
pantiesly
pantiess
panty
pantyed
pantyer
pantyes
pantying
pantyly
pantys
pastie
pastieed
pastieer
pastiees
pastieing
pastiely
pasties
pasty
pastyed
pastyer
pastyes
pastying
pastyly
pastys
pecker
peckered
peckerer
peckeres
peckering
peckerly
peckers
pedo
pedoed
pedoer
pedoes
pedoing
pedoly
pedophile
pedophileed
pedophileer
pedophilees
pedophileing
pedophilely
pedophiles
pedophilia
pedophiliac
pedophiliaced
pedophiliacer
pedophiliaces
pedophiliacing
pedophiliacly
pedophiliacs
pedophiliaed
pedophiliaer
pedophiliaes
pedophiliaing
pedophilialy
pedophilias
pedos
penial
penialed
penialer
peniales
penialing
penially
penials
penile
penileed
penileer
penilees
penileing
penilely
peniles
penis
penised
peniser
penises
penising
penisly
peniss
perversion
perversioned
perversioner
perversiones
perversioning
perversionly
perversions
peyote
peyoteed
peyoteer
peyotees
peyoteing
peyotely
peyotes
phuck
phucked
phucker
phuckes
phucking
phuckly
phucks
pillowbiter
pillowbitered
pillowbiterer
pillowbiteres
pillowbitering
pillowbiterly
pillowbiters
pimp
pimped
pimper
pimpes
pimping
pimply
pimps
pinko
pinkoed
pinkoer
pinkoes
pinkoing
pinkoly
pinkos
pissed
pisseded
pisseder
pissedes
pisseding
pissedly
pisseds
pisser
pisses
pissing
pissly
pissoff
pissoffed
pissoffer
pissoffes
pissoffing
pissoffly
pissoffs
pisss
polack
polacked
polacker
polackes
polacking
polackly
polacks
pollock
pollocked
pollocker
pollockes
pollocking
pollockly
pollocks
poon
pooned
pooner
poones
pooning
poonly
poons
poontang
poontanged
poontanger
poontanges
poontanging
poontangly
poontangs
porn
porned
porner
pornes
porning
pornly
porno
pornoed
pornoer
pornoes
pornography
pornographyed
pornographyer
pornographyes
pornographying
pornographyly
pornographys
pornoing
pornoly
pornos
porns
prick
pricked
pricker
prickes
pricking
prickly
pricks
prig
priged
priger
priges
priging
prigly
prigs
prostitute
prostituteed
prostituteer
prostitutees
prostituteing
prostitutely
prostitutes
prude
prudeed
prudeer
prudees
prudeing
prudely
prudes
punkass
punkassed
punkasser
punkasses
punkassing
punkassly
punkasss
punky
punkyed
punkyer
punkyes
punkying
punkyly
punkys
puss
pussed
pusser
pusses
pussies
pussiesed
pussieser
pussieses
pussiesing
pussiesly
pussiess
pussing
pussly
pusss
pussy
pussyed
pussyer
pussyes
pussying
pussyly
pussypounder
pussypoundered
pussypounderer
pussypounderes
pussypoundering
pussypounderly
pussypounders
pussys
puto
putoed
putoer
putoes
putoing
putoly
putos
queaf
queafed
queafer
queafes
queafing
queafly
queafs
queef
queefed
queefer
queefes
queefing
queefly
queefs
queer
queered
queerer
queeres
queering
queerly
queero
queeroed
queeroer
queeroes
queeroing
queeroly
queeros
queers
queersed
queerser
queerses
queersing
queersly
queerss
quicky
quickyed
quickyer
quickyes
quickying
quickyly
quickys
quim
quimed
quimer
quimes
quiming
quimly
quims
racy
racyed
racyer
racyes
racying
racyly
racys
rape
raped
rapeded
rapeder
rapedes
rapeding
rapedly
rapeds
rapeed
rapeer
rapees
rapeing
rapely
raper
rapered
raperer
raperes
rapering
raperly
rapers
rapes
rapist
rapisted
rapister
rapistes
rapisting
rapistly
rapists
raunch
raunched
rauncher
raunches
raunching
raunchly
raunchs
rectus
rectused
rectuser
rectuses
rectusing
rectusly
rectuss
reefer
reefered
reeferer
reeferes
reefering
reeferly
reefers
reetard
reetarded
reetarder
reetardes
reetarding
reetardly
reetards
reich
reiched
reicher
reiches
reiching
reichly
reichs
retard
retarded
retardeded
retardeder
retardedes
retardeding
retardedly
retardeds
retarder
retardes
retarding
retardly
retards
rimjob
rimjobed
rimjober
rimjobes
rimjobing
rimjobly
rimjobs
ritard
ritarded
ritarder
ritardes
ritarding
ritardly
ritards
rtard
rtarded
rtarder
rtardes
rtarding
rtardly
rtards
rum
rumed
rumer
rumes
ruming
rumly
rump
rumped
rumper
rumpes
rumping
rumply
rumprammer
rumprammered
rumprammerer
rumprammeres
rumprammering
rumprammerly
rumprammers
rumps
rums
ruski
ruskied
ruskier
ruskies
ruskiing
ruskily
ruskis
sadism
sadismed
sadismer
sadismes
sadisming
sadismly
sadisms
sadist
sadisted
sadister
sadistes
sadisting
sadistly
sadists
scag
scaged
scager
scages
scaging
scagly
scags
scantily
scantilyed
scantilyer
scantilyes
scantilying
scantilyly
scantilys
schlong
schlonged
schlonger
schlonges
schlonging
schlongly
schlongs
scrog
scroged
scroger
scroges
scroging
scrogly
scrogs
scrot
scrote
scroted
scroteed
scroteer
scrotees
scroteing
scrotely
scroter
scrotes
scroting
scrotly
scrots
scrotum
scrotumed
scrotumer
scrotumes
scrotuming
scrotumly
scrotums
scrud
scruded
scruder
scrudes
scruding
scrudly
scruds
scum
scumed
scumer
scumes
scuming
scumly
scums
seaman
seamaned
seamaner
seamanes
seamaning
seamanly
seamans
seamen
seamened
seamener
seamenes
seamening
seamenly
seamens
seduceed
seduceer
seducees
seduceing
seducely
seduces
semen
semened
semener
semenes
semening
semenly
semens
shamedame
shamedameed
shamedameer
shamedamees
shamedameing
shamedamely
shamedames
shit
shite
shiteater
shiteatered
shiteaterer
shiteateres
shiteatering
shiteaterly
shiteaters
shited
shiteed
shiteer
shitees
shiteing
shitely
shiter
shites
shitface
shitfaceed
shitfaceer
shitfacees
shitfaceing
shitfacely
shitfaces
shithead
shitheaded
shitheader
shitheades
shitheading
shitheadly
shitheads
shithole
shitholeed
shitholeer
shitholees
shitholeing
shitholely
shitholes
shithouse
shithouseed
shithouseer
shithousees
shithouseing
shithousely
shithouses
shiting
shitly
shits
shitsed
shitser
shitses
shitsing
shitsly
shitss
shitt
shitted
shitteded
shitteder
shittedes
shitteding
shittedly
shitteds
shitter
shittered
shitterer
shitteres
shittering
shitterly
shitters
shittes
shitting
shittly
shitts
shitty
shittyed
shittyer
shittyes
shittying
shittyly
shittys
shiz
shized
shizer
shizes
shizing
shizly
shizs
shooted
shooter
shootes
shooting
shootly
shoots
sissy
sissyed
sissyer
sissyes
sissying
sissyly
sissys
skag
skaged
skager
skages
skaging
skagly
skags
skank
skanked
skanker
skankes
skanking
skankly
skanks
slave
slaveed
slaveer
slavees
slaveing
slavely
slaves
sleaze
sleazeed
sleazeer
sleazees
sleazeing
sleazely
sleazes
sleazy
sleazyed
sleazyer
sleazyes
sleazying
sleazyly
sleazys
slut
slutdumper
slutdumpered
slutdumperer
slutdumperes
slutdumpering
slutdumperly
slutdumpers
sluted
sluter
slutes
sluting
slutkiss
slutkissed
slutkisser
slutkisses
slutkissing
slutkissly
slutkisss
slutly
sluts
slutsed
slutser
slutses
slutsing
slutsly
slutss
smegma
smegmaed
smegmaer
smegmaes
smegmaing
smegmaly
smegmas
smut
smuted
smuter
smutes
smuting
smutly
smuts
smutty
smuttyed
smuttyer
smuttyes
smuttying
smuttyly
smuttys
snatch
snatched
snatcher
snatches
snatching
snatchly
snatchs
sniper
snipered
sniperer
sniperes
snipering
sniperly
snipers
snort
snorted
snorter
snortes
snorting
snortly
snorts
snuff
snuffed
snuffer
snuffes
snuffing
snuffly
snuffs
sodom
sodomed
sodomer
sodomes
sodoming
sodomly
sodoms
spic
spiced
spicer
spices
spicing
spick
spicked
spicker
spickes
spicking
spickly
spicks
spicly
spics
spik
spoof
spoofed
spoofer
spoofes
spoofing
spoofly
spoofs
spooge
spoogeed
spoogeer
spoogees
spoogeing
spoogely
spooges
spunk
spunked
spunker
spunkes
spunking
spunkly
spunks
steamyed
steamyer
steamyes
steamying
steamyly
steamys
stfu
stfued
stfuer
stfues
stfuing
stfuly
stfus
stiffy
stiffyed
stiffyer
stiffyes
stiffying
stiffyly
stiffys
stoneded
stoneder
stonedes
stoneding
stonedly
stoneds
stupided
stupider
stupides
stupiding
stupidly
stupids
suckeded
suckeder
suckedes
suckeding
suckedly
suckeds
sucker
suckes
sucking
suckinged
suckinger
suckinges
suckinging
suckingly
suckings
suckly
sucks
sumofabiatch
sumofabiatched
sumofabiatcher
sumofabiatches
sumofabiatching
sumofabiatchly
sumofabiatchs
tard
tarded
tarder
tardes
tarding
tardly
tards
tawdry
tawdryed
tawdryer
tawdryes
tawdrying
tawdryly
tawdrys
teabagging
teabagginged
teabagginger
teabagginges
teabagginging
teabaggingly
teabaggings
terd
terded
terder
terdes
terding
terdly
terds
teste
testee
testeed
testeeed
testeeer
testeees
testeeing
testeely
testeer
testees
testeing
testely
testes
testesed
testeser
testeses
testesing
testesly
testess
testicle
testicleed
testicleer
testiclees
testicleing
testiclely
testicles
testis
testised
testiser
testises
testising
testisly
testiss
thrusted
thruster
thrustes
thrusting
thrustly
thrusts
thug
thuged
thuger
thuges
thuging
thugly
thugs
tinkle
tinkleed
tinkleer
tinklees
tinkleing
tinklely
tinkles
tit
tited
titer
tites
titfuck
titfucked
titfucker
titfuckes
titfucking
titfuckly
titfucks
titi
titied
titier
tities
titiing
titily
titing
titis
titly
tits
titsed
titser
titses
titsing
titsly
titss
tittiefucker
tittiefuckered
tittiefuckerer
tittiefuckeres
tittiefuckering
tittiefuckerly
tittiefuckers
titties
tittiesed
tittieser
tittieses
tittiesing
tittiesly
tittiess
titty
tittyed
tittyer
tittyes
tittyfuck
tittyfucked
tittyfucker
tittyfuckered
tittyfuckerer
tittyfuckeres
tittyfuckering
tittyfuckerly
tittyfuckers
tittyfuckes
tittyfucking
tittyfuckly
tittyfucks
tittying
tittyly
tittys
toke
tokeed
tokeer
tokees
tokeing
tokely
tokes
toots
tootsed
tootser
tootses
tootsing
tootsly
tootss
tramp
tramped
tramper
trampes
tramping
tramply
tramps
transsexualed
transsexualer
transsexuales
transsexualing
transsexually
transsexuals
trashy
trashyed
trashyer
trashyes
trashying
trashyly
trashys
tubgirl
tubgirled
tubgirler
tubgirles
tubgirling
tubgirlly
tubgirls
turd
turded
turder
turdes
turding
turdly
turds
tush
tushed
tusher
tushes
tushing
tushly
tushs
twat
twated
twater
twates
twating
twatly
twats
twatsed
twatser
twatses
twatsing
twatsly
twatss
undies
undiesed
undieser
undieses
undiesing
undiesly
undiess
unweded
unweder
unwedes
unweding
unwedly
unweds
uzi
uzied
uzier
uzies
uziing
uzily
uzis
vag
vaged
vager
vages
vaging
vagly
vags
valium
valiumed
valiumer
valiumes
valiuming
valiumly
valiums
venous
virgined
virginer
virgines
virgining
virginly
virgins
vixen
vixened
vixener
vixenes
vixening
vixenly
vixens
vodkaed
vodkaer
vodkaes
vodkaing
vodkaly
vodkas
voyeur
voyeured
voyeurer
voyeures
voyeuring
voyeurly
voyeurs
vulgar
vulgared
vulgarer
vulgares
vulgaring
vulgarly
vulgars
wang
wanged
wanger
wanges
wanging
wangly
wangs
wank
wanked
wanker
wankered
wankerer
wankeres
wankering
wankerly
wankers
wankes
wanking
wankly
wanks
wazoo
wazooed
wazooer
wazooes
wazooing
wazooly
wazoos
wedgie
wedgieed
wedgieer
wedgiees
wedgieing
wedgiely
wedgies
weeded
weeder
weedes
weeding
weedly
weeds
weenie
weenieed
weenieer
weeniees
weenieing
weeniely
weenies
weewee
weeweeed
weeweeer
weeweees
weeweeing
weeweely
weewees
weiner
weinered
weinerer
weineres
weinering
weinerly
weiners
weirdo
weirdoed
weirdoer
weirdoes
weirdoing
weirdoly
weirdos
wench
wenched
wencher
wenches
wenching
wenchly
wenchs
wetback
wetbacked
wetbacker
wetbackes
wetbacking
wetbackly
wetbacks
whitey
whiteyed
whiteyer
whiteyes
whiteying
whiteyly
whiteys
whiz
whized
whizer
whizes
whizing
whizly
whizs
whoralicious
whoralicioused
whoraliciouser
whoraliciouses
whoraliciousing
whoraliciously
whoraliciouss
whore
whorealicious
whorealicioused
whorealiciouser
whorealiciouses
whorealiciousing
whorealiciously
whorealiciouss
whored
whoreded
whoreder
whoredes
whoreding
whoredly
whoreds
whoreed
whoreer
whorees
whoreface
whorefaceed
whorefaceer
whorefacees
whorefaceing
whorefacely
whorefaces
whorehopper
whorehoppered
whorehopperer
whorehopperes
whorehoppering
whorehopperly
whorehoppers
whorehouse
whorehouseed
whorehouseer
whorehousees
whorehouseing
whorehousely
whorehouses
whoreing
whorely
whores
whoresed
whoreser
whoreses
whoresing
whoresly
whoress
whoring
whoringed
whoringer
whoringes
whoringing
whoringly
whorings
wigger
wiggered
wiggerer
wiggeres
wiggering
wiggerly
wiggers
woody
woodyed
woodyer
woodyes
woodying
woodyly
woodys
wop
woped
woper
wopes
woping
woply
wops
wtf
wtfed
wtfer
wtfes
wtfing
wtfly
wtfs
xxx
xxxed
xxxer
xxxes
xxxing
xxxly
xxxs
yeasty
yeastyed
yeastyer
yeastyes
yeastying
yeastyly
yeastys
yobbo
yobboed
yobboer
yobboes
yobboing
yobboly
yobbos
zoophile
zoophileed
zoophileer
zoophilees
zoophileing
zoophilely
zoophiles
anal
ass
ass lick
balls
ballsac
bisexual
bleach
causas
cheap
cost of miracles
cunt
display network stats
fart
fda and death
fda AND warn
fda AND warning
fda AND warns
feom
fuck
gfc
humira AND expensive
illegal
madvocate
masturbation
nuccitelli
overdose
porn
shit
snort
texarkana
direct\-acting antivirals
assistance
ombitasvir
support path
harvoni
abbvie
direct-acting antivirals
paritaprevir
advocacy
ledipasvir
vpak
ritonavir with dasabuvir
program
gilead
greedy
financial
needy
fake-ovir
viekira pak
v pak
sofosbuvir
support
oasis
discount
dasabuvir
protest
ritonavir
section[contains(@class, 'nav-hidden')]
footer[@id='footer']
div[contains(@class, 'pane-pub-article-cleveland-clinic')]
div[contains(@class, 'pane-pub-home-cleveland-clinic')]
div[contains(@class, 'pane-pub-topic-cleveland-clinic')]
div[contains(@class, 'panel-panel-inner')]
div[contains(@class, 'pane-node-field-article-topics')]
section[contains(@class, 'footer-nav-section-wrapper')]
Pharmacogenomics for the primary care provider: Why should we care?
Since the human genome was sequenced in 2000, the American public has continued to hold hope that our growing understanding of genetics will revolutionize the practice of medicine.
One way genetics promises to improve the quality and value of health care is in personalized medicine, by helping us tailor treatment to a person’s individual genetic makeup. One such approach is called pharmacogenomics.
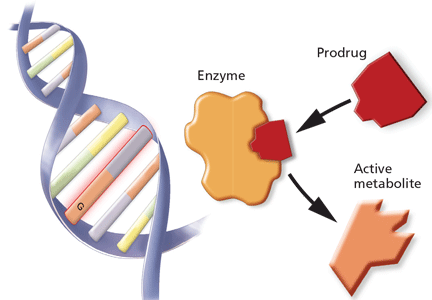
Pharmacogenomics uses knowledge of a person’s genetics to understand how a particular drug will work, or not work, in his or her body. For instance, some people might carry genes that make them more sensitive than average to a drug, and therefore they would require a lower dose. Others might have genes that make them resistant to the drug, meaning the drug is ineffective unless they receive a higher dose.
Adverse drug reactions are a leading cause of death in hospitalized patients in the United States and are responsible for billions of dollars in health care costs.1,2 Our current practice of prescribing for adult patients is largely trial-and-error, with the same dose given to all patients, in many cases with little regard even to sex, height, or weight.
Pharmacogenomics promises to change this way of prescribing to a customized approach that uses genetic information to predict an individual’s response to medications. It is one piece of an overall initiative to personalize patient care based on the patient’s individual characteristics and preferences.
OVERCOMING BARRIERS TO USING PHARMACOGENOMICS IN PRACTICE
If personalized medicine has promised to improve the quality and value of health care for our patients, why have we been so slow to adopt this information in clinical practice?
The usual barriers to clinical adoption certainly exist. We need further studies to determine whether genetic-based prescribing is truly valid, and for which patient populations. We need to determine whether this approach is cost-effective and better than the current standard of care. We need to work on payment options.
However, one of the largest barriers for busy primary care physicians is the lack of time to keep up with new information. Many practicing physicians were taught little about formal genetics in medical school. The body of scientific literature on pharmacogenomics is fragmented, and it crosses disease states and specialties, making it difficult to unite. Given the breadth of diseases treated and drugs prescribed by primary care physicians, it is unrealistic for most to keep track of the vast body of literature of pharmacogenomic testing and to decipher how to apply this to clinical practice.
In this issue of the Journal, Kitzmiller et al3 provide one solution to this problem, giving an overview of pharmacogenomic applications that might be pertinent to practicing physicians. However, as we try to make pharmacogenomics accessible to busy physicians, we need other solutions to integrate pharmacogenomic information efficiently into the clinical work flow. One approach might be to build pharmacogenomics into the electronic medical record. We can also store the integrated information in research databases and provide clinical recommendations on Internet sites such as www.pharmgkb.org, and we can develop applications to run on cell phones and iPads.
QUESTIONS REMAIN
Kitzmiller et al discuss an important step in this process, highlighting several key questions:
Should we seek genetics-based information to personalize drug selection? Based on the information presented in the literature and in the Kitzmiller paper, there may be circumstances when it is appropriate to consider doing so. While the evidence is not yet compelling to order these tests on a regular basis in clinical practice, this information might be helpful in some situations, such as for patients who have had adverse effects from minimal doses of antidepressants.
For now, clinicians should not abandon their current practice of personalizing patient care on the basis of personal, cultural, and economic preferences. Rather, they should consider pharmacogenomic information an additional piece of information when selecting drug therapy. We should also encourage health care systems and interested providers to be early adopters and to study how their outcomes compare with the standard of care.
Once we have this information, what is our obligation to use it? An increasing number of patients already have genetic information in their health record, either ordered by or provided to their physicians. However, there is little in the scientific literature to guide us in this arena.
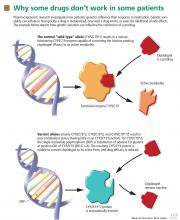
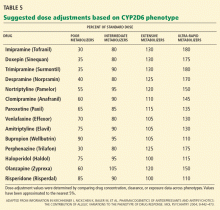
Yet most of us would agree that if we have information (genetic or otherwise) that can help to select a drug type or dose or reduce adverse events or costs, we should consider this information in our decision-making. Several circumstances are documented in this paper and in the literature in which prior knowledge about drug metabolism can help in selecting a dose of medication. One example would be the 50% recommended reduction in tricyclic antidepressant dose if the patient is a CYP2D6 poor metabolizer.4
MOVING FORWARD AS A TEAM
In summary, Kitzmiller et al bring to light the promise and the uncertainties that currently exist in the field of pharmacogenomics. While it is unclear if we should incorporate pharmacogenomic tests into standard medical practice at this time, it is clear that this information is becoming more readily available, whether or not we have requested it. Some would argue that, once we have the information, we have an obligation to use it, just as we use other information in our clinical decision-making. This means we need to develop tools and resources to help practitioners evaluate pharmacogenomic data and incorporate it into clinical care in an efficient manner.
The authors also highlight the need for more education about drug metabolism in general, and they cite several instances in which knowledge of drug interactions and metabolism can clearly influence decision-making. An example is paroxetine (Paxil) inhibition of tamoxifen (Nolvadex).5
Lastly, regardless of our personal feelings about the clinical usefulness of genetic testing in large populations, we need to work together to determine clinical utility and validity and to develop efficient ways to put into practice findings that could affect patient care. As we move forward, we need to work as a team, utilizing our clinical partners—pharmacists, pharmacologists, metabolism and health information technology experts, and medical geneticists. Working as a team, pooling our resources and tools, we move closer to providing world-class personalized health care.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998; 279:1200–1205.
- Field TS, Gilman BH, Subramanian S, Fuller JC, Bates DW, Gurwitz JH. The costs associated with adverse drug events among older adults in the ambulatory setting. Med Care 2005; 43:1171–1176.
- Kitzmiller JP, Groen DK, Phelps MA, Sadee W. Pharmacogenomic testing: relevance in medical practice. Why drugs work in some patients but not others. Cleve Clin J Med 2011; 78:243–257.
- Thuerauf N, Lunkenheimer J. The impact of the CYP2D6-polymorphism on dose recommendations for current antidepressants. Eur Arch Psychiatry Clin Neurosci 2006; 256:287–293.
- Schwarz EB, McNamara M, Miller RG, Walsh JM. Update in women’s health for the general internist. J Gen Intern Med201; 26:207–213.
Since the human genome was sequenced in 2000, the American public has continued to hold hope that our growing understanding of genetics will revolutionize the practice of medicine.
One way genetics promises to improve the quality and value of health care is in personalized medicine, by helping us tailor treatment to a person’s individual genetic makeup. One such approach is called pharmacogenomics.
Pharmacogenomics uses knowledge of a person’s genetics to understand how a particular drug will work, or not work, in his or her body. For instance, some people might carry genes that make them more sensitive than average to a drug, and therefore they would require a lower dose. Others might have genes that make them resistant to the drug, meaning the drug is ineffective unless they receive a higher dose.
Adverse drug reactions are a leading cause of death in hospitalized patients in the United States and are responsible for billions of dollars in health care costs.1,2 Our current practice of prescribing for adult patients is largely trial-and-error, with the same dose given to all patients, in many cases with little regard even to sex, height, or weight.
Pharmacogenomics promises to change this way of prescribing to a customized approach that uses genetic information to predict an individual’s response to medications. It is one piece of an overall initiative to personalize patient care based on the patient’s individual characteristics and preferences.
OVERCOMING BARRIERS TO USING PHARMACOGENOMICS IN PRACTICE
If personalized medicine has promised to improve the quality and value of health care for our patients, why have we been so slow to adopt this information in clinical practice?
The usual barriers to clinical adoption certainly exist. We need further studies to determine whether genetic-based prescribing is truly valid, and for which patient populations. We need to determine whether this approach is cost-effective and better than the current standard of care. We need to work on payment options.
However, one of the largest barriers for busy primary care physicians is the lack of time to keep up with new information. Many practicing physicians were taught little about formal genetics in medical school. The body of scientific literature on pharmacogenomics is fragmented, and it crosses disease states and specialties, making it difficult to unite. Given the breadth of diseases treated and drugs prescribed by primary care physicians, it is unrealistic for most to keep track of the vast body of literature of pharmacogenomic testing and to decipher how to apply this to clinical practice.
In this issue of the Journal, Kitzmiller et al3 provide one solution to this problem, giving an overview of pharmacogenomic applications that might be pertinent to practicing physicians. However, as we try to make pharmacogenomics accessible to busy physicians, we need other solutions to integrate pharmacogenomic information efficiently into the clinical work flow. One approach might be to build pharmacogenomics into the electronic medical record. We can also store the integrated information in research databases and provide clinical recommendations on Internet sites such as www.pharmgkb.org, and we can develop applications to run on cell phones and iPads.
QUESTIONS REMAIN
Kitzmiller et al discuss an important step in this process, highlighting several key questions:
Should we seek genetics-based information to personalize drug selection? Based on the information presented in the literature and in the Kitzmiller paper, there may be circumstances when it is appropriate to consider doing so. While the evidence is not yet compelling to order these tests on a regular basis in clinical practice, this information might be helpful in some situations, such as for patients who have had adverse effects from minimal doses of antidepressants.
For now, clinicians should not abandon their current practice of personalizing patient care on the basis of personal, cultural, and economic preferences. Rather, they should consider pharmacogenomic information an additional piece of information when selecting drug therapy. We should also encourage health care systems and interested providers to be early adopters and to study how their outcomes compare with the standard of care.
Once we have this information, what is our obligation to use it? An increasing number of patients already have genetic information in their health record, either ordered by or provided to their physicians. However, there is little in the scientific literature to guide us in this arena.
Yet most of us would agree that if we have information (genetic or otherwise) that can help to select a drug type or dose or reduce adverse events or costs, we should consider this information in our decision-making. Several circumstances are documented in this paper and in the literature in which prior knowledge about drug metabolism can help in selecting a dose of medication. One example would be the 50% recommended reduction in tricyclic antidepressant dose if the patient is a CYP2D6 poor metabolizer.4
MOVING FORWARD AS A TEAM
In summary, Kitzmiller et al bring to light the promise and the uncertainties that currently exist in the field of pharmacogenomics. While it is unclear if we should incorporate pharmacogenomic tests into standard medical practice at this time, it is clear that this information is becoming more readily available, whether or not we have requested it. Some would argue that, once we have the information, we have an obligation to use it, just as we use other information in our clinical decision-making. This means we need to develop tools and resources to help practitioners evaluate pharmacogenomic data and incorporate it into clinical care in an efficient manner.
The authors also highlight the need for more education about drug metabolism in general, and they cite several instances in which knowledge of drug interactions and metabolism can clearly influence decision-making. An example is paroxetine (Paxil) inhibition of tamoxifen (Nolvadex).5
Lastly, regardless of our personal feelings about the clinical usefulness of genetic testing in large populations, we need to work together to determine clinical utility and validity and to develop efficient ways to put into practice findings that could affect patient care. As we move forward, we need to work as a team, utilizing our clinical partners—pharmacists, pharmacologists, metabolism and health information technology experts, and medical geneticists. Working as a team, pooling our resources and tools, we move closer to providing world-class personalized health care.
Since the human genome was sequenced in 2000, the American public has continued to hold hope that our growing understanding of genetics will revolutionize the practice of medicine.
One way genetics promises to improve the quality and value of health care is in personalized medicine, by helping us tailor treatment to a person’s individual genetic makeup. One such approach is called pharmacogenomics.
Pharmacogenomics uses knowledge of a person’s genetics to understand how a particular drug will work, or not work, in his or her body. For instance, some people might carry genes that make them more sensitive than average to a drug, and therefore they would require a lower dose. Others might have genes that make them resistant to the drug, meaning the drug is ineffective unless they receive a higher dose.
Adverse drug reactions are a leading cause of death in hospitalized patients in the United States and are responsible for billions of dollars in health care costs.1,2 Our current practice of prescribing for adult patients is largely trial-and-error, with the same dose given to all patients, in many cases with little regard even to sex, height, or weight.
Pharmacogenomics promises to change this way of prescribing to a customized approach that uses genetic information to predict an individual’s response to medications. It is one piece of an overall initiative to personalize patient care based on the patient’s individual characteristics and preferences.
OVERCOMING BARRIERS TO USING PHARMACOGENOMICS IN PRACTICE
If personalized medicine has promised to improve the quality and value of health care for our patients, why have we been so slow to adopt this information in clinical practice?
The usual barriers to clinical adoption certainly exist. We need further studies to determine whether genetic-based prescribing is truly valid, and for which patient populations. We need to determine whether this approach is cost-effective and better than the current standard of care. We need to work on payment options.
However, one of the largest barriers for busy primary care physicians is the lack of time to keep up with new information. Many practicing physicians were taught little about formal genetics in medical school. The body of scientific literature on pharmacogenomics is fragmented, and it crosses disease states and specialties, making it difficult to unite. Given the breadth of diseases treated and drugs prescribed by primary care physicians, it is unrealistic for most to keep track of the vast body of literature of pharmacogenomic testing and to decipher how to apply this to clinical practice.
In this issue of the Journal, Kitzmiller et al3 provide one solution to this problem, giving an overview of pharmacogenomic applications that might be pertinent to practicing physicians. However, as we try to make pharmacogenomics accessible to busy physicians, we need other solutions to integrate pharmacogenomic information efficiently into the clinical work flow. One approach might be to build pharmacogenomics into the electronic medical record. We can also store the integrated information in research databases and provide clinical recommendations on Internet sites such as www.pharmgkb.org, and we can develop applications to run on cell phones and iPads.
QUESTIONS REMAIN
Kitzmiller et al discuss an important step in this process, highlighting several key questions:
Should we seek genetics-based information to personalize drug selection? Based on the information presented in the literature and in the Kitzmiller paper, there may be circumstances when it is appropriate to consider doing so. While the evidence is not yet compelling to order these tests on a regular basis in clinical practice, this information might be helpful in some situations, such as for patients who have had adverse effects from minimal doses of antidepressants.
For now, clinicians should not abandon their current practice of personalizing patient care on the basis of personal, cultural, and economic preferences. Rather, they should consider pharmacogenomic information an additional piece of information when selecting drug therapy. We should also encourage health care systems and interested providers to be early adopters and to study how their outcomes compare with the standard of care.
Once we have this information, what is our obligation to use it? An increasing number of patients already have genetic information in their health record, either ordered by or provided to their physicians. However, there is little in the scientific literature to guide us in this arena.
Yet most of us would agree that if we have information (genetic or otherwise) that can help to select a drug type or dose or reduce adverse events or costs, we should consider this information in our decision-making. Several circumstances are documented in this paper and in the literature in which prior knowledge about drug metabolism can help in selecting a dose of medication. One example would be the 50% recommended reduction in tricyclic antidepressant dose if the patient is a CYP2D6 poor metabolizer.4
MOVING FORWARD AS A TEAM
In summary, Kitzmiller et al bring to light the promise and the uncertainties that currently exist in the field of pharmacogenomics. While it is unclear if we should incorporate pharmacogenomic tests into standard medical practice at this time, it is clear that this information is becoming more readily available, whether or not we have requested it. Some would argue that, once we have the information, we have an obligation to use it, just as we use other information in our clinical decision-making. This means we need to develop tools and resources to help practitioners evaluate pharmacogenomic data and incorporate it into clinical care in an efficient manner.
The authors also highlight the need for more education about drug metabolism in general, and they cite several instances in which knowledge of drug interactions and metabolism can clearly influence decision-making. An example is paroxetine (Paxil) inhibition of tamoxifen (Nolvadex).5
Lastly, regardless of our personal feelings about the clinical usefulness of genetic testing in large populations, we need to work together to determine clinical utility and validity and to develop efficient ways to put into practice findings that could affect patient care. As we move forward, we need to work as a team, utilizing our clinical partners—pharmacists, pharmacologists, metabolism and health information technology experts, and medical geneticists. Working as a team, pooling our resources and tools, we move closer to providing world-class personalized health care.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998; 279:1200–1205.
- Field TS, Gilman BH, Subramanian S, Fuller JC, Bates DW, Gurwitz JH. The costs associated with adverse drug events among older adults in the ambulatory setting. Med Care 2005; 43:1171–1176.
- Kitzmiller JP, Groen DK, Phelps MA, Sadee W. Pharmacogenomic testing: relevance in medical practice. Why drugs work in some patients but not others. Cleve Clin J Med 2011; 78:243–257.
- Thuerauf N, Lunkenheimer J. The impact of the CYP2D6-polymorphism on dose recommendations for current antidepressants. Eur Arch Psychiatry Clin Neurosci 2006; 256:287–293.
- Schwarz EB, McNamara M, Miller RG, Walsh JM. Update in women’s health for the general internist. J Gen Intern Med201; 26:207–213.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998; 279:1200–1205.
- Field TS, Gilman BH, Subramanian S, Fuller JC, Bates DW, Gurwitz JH. The costs associated with adverse drug events among older adults in the ambulatory setting. Med Care 2005; 43:1171–1176.
- Kitzmiller JP, Groen DK, Phelps MA, Sadee W. Pharmacogenomic testing: relevance in medical practice. Why drugs work in some patients but not others. Cleve Clin J Med 2011; 78:243–257.
- Thuerauf N, Lunkenheimer J. The impact of the CYP2D6-polymorphism on dose recommendations for current antidepressants. Eur Arch Psychiatry Clin Neurosci 2006; 256:287–293.
- Schwarz EB, McNamara M, Miller RG, Walsh JM. Update in women’s health for the general internist. J Gen Intern Med201; 26:207–213.
Pharmacogenomic testing: Relevance in medical practice
In many patients, certain drugs do not work as well as expected, whereas in other patients they cause toxic effects, even at lower doses. For some patients, the reason may be genetic.
Sizeable minorities of the population carry genetic variants—polymorphisms— that affect their response to various drugs. Thanks to genetic research, our understanding of the variability of drug response has advanced markedly in the last decade. Many relevant polymorphisms have been identified, and tests for some of them are available.
Armed with the knowledge of their patients’ genetic status, physicians could predict their response to certain drugs, leading to better efficacy, fewer adverse drug reactions, and a better cost-benefit ratio.
The possible impact is substantial, since more than half of the drugs most commonly involved in adverse drug reactions are metabolized by polymorphic enzymes.1 Adverse drug reactions remain a significant detriment to public health, having a substantial impact on rates of morbidity and death and on healthcare costs.2–8 In the United States, adverse drug reactions are a leading cause of death in hospitalized patients4 and are annually responsible for hundreds of thousands of deaths and hundreds of billions of dollars in added costs.2,4,6–8
In the meantime, physicians can educate their patients and promote efforts to incorporate genomic information into standard clinical decision-making.
This article offers an overview of pharmacogenomic testing, discussing implications and limitations of a few validated tests. Specifically, we will discuss testing that is relevant when using warfarin (Coumadin), clopidogrel (Plavix), statins, tamoxifen (Nolvadex), codeine, and psychotropic medications, as well as the future role of pharmacogenomic testing in medicine.
WHAT IS PHARMACOGENOMICS?
Pharmacogenomics is the study of how genetic factors relate to interindividual variability of drug response.
Many clinicians may not be familiar with the background and terminology used in the pharmacogenomic literature. Below, a brief review of the terminology is followed by a schematic describing the various stages of research involved in pharmacogenomics and the advancement of a test into standard practice.
The review and schematic may be helpful for evaluating the clinical significance of pharmacogenomics-related articles.
From genotype to phenotype
Genotype refers to the coding sequence of DNA base pairs for a particular gene, and phenotype (eg, disease or drug response) refers to a trait resulting from the protein product encoded by the gene. The name of a gene often refers to its protein product and is italicized (eg, the CYP3A4 gene encodes for the CYP3A4 enzyme).
Two alleles per autosomal gene (one paternal and one maternal) form the genotype. Heterozygotes possess two different alleles, and homozygotes possess two of the same alleles. The most common allele in a population is referred to as the wild type, and allele frequencies can vary greatly in different populations.9
Most sequence variations are single nucleotide polymorphisms (SNPs, pronounced “snips”), a single DNA base pair substitution that may result in a different gene product. SNPs can be classified as structural RNA polymorphisms (srSNPs), regulatory polymorphisms (rSNPs), or polymorphisms in coding regions (cSNPs)10: srSNPs alter mRNA processing and translation, rSNPs alter transcription, and cSNPs alter protein sequence and function.
Recently, genetic associations with a phenotype have been done on a large scale, with millions of SNPs measured in each of many subjects. This approach, called a genomewide association study or GWAS, has revealed countless candidate genes for clinical traits, but only a few have resulted in a practical clinical application. SNPs may by themselves exert a pharmacokinetic effect (ie, how the body processes the drug), a pharmacodynamic effect (ie, how the drug affects the body), or both, or they may act in concert with other genetic factors. Pharmacodynamic effects can result from a pharmacokinetic effect or can result from variations in a pharmacologic target.
Establishing a genotype-phenotype association can involve clinical studies, animal transgenic studies, or molecular and cellular functional assays.
Clinical applications are emerging
WARFARIN: IMPORTANCE OF CYP2C9, VKORC1
Warfarin is used for the long-term treatment and prevention of thromboembolic events.
This drug has a narrow therapeutic window and shows substantial interpatient dose variability. The start of warfarin therapy is associated with one of the highest rates of adverse events and emergency room visits of any single drug.12 More than 2 million patients start warfarin each year in the United States alone,13 and about 20% of them are hospitalized within the first 6 months because of bleeding due to overanticoagulation.14
The findings from a recent study suggest that pharmacogenomic testing may eventually allow more patients to safely benefit from warfarin therapy. In this large, nationwide, prospective study, hospitalization rates were 30% lower when pharmacogenomic testing was used.14 However, no reduction was seen in the time needed to reach the target international normalized ratio (INR) or in the need for INR checks at 6 months. Furthermore, this study used historical control data, and some or all of the reduction in hospitalization rates may be attributed to more frequent INR checks in the patients who underwent testing than in the historical control group.
A relationship between warfarin dose requirements and the genetic status of CYP2C9, which encodes a major drug-metabolizing enzyme, has been demonstrated in retrospective and prospective studies.15–17
S-warfarin is metabolized by CYP2C9, which is polymorphic
Warfarin contains equal amounts of two isomers, designated S and R. S-warfarin, which is more potent, is metabolized principally by CYP2C9, while R-warfarin is metabolized by CYP1A2, CYP2C19, and CYP3A4.
People who possess two copies of the wild type CYP2C9 gene CYP2C9*1 metabolize warfarin very well and so are called “extensive warfarin metabolizers.” Carriers of the allelic variants CYP2C9*2 and CYP2C9*3 (which have point mutations in exons 3 and 7 of CYP2C9, respectively), have less capacity. Compared with those who are homozygous for the wild-type gene, homozygous carriers of CYP2C9* 3 clear S-warfarin at a rate that is 90% lower, and those with the CYP2C9*1/*3, CYP2C9* 1/*2, CYP2C9*2/*2, or CYP2C9*2/*3 genotypes clear it at a rate 50% to 75% lower. A meta-analysis of 12 studies found that the CYP2C9 genotype accounted for 12% of the interindividual variability of warfarin dose requirements.18
About 8% of whites carry at least one copy of CYP2C9*2, as do 1% of African Americans; the allele is rare in Asian populations. The frequency of CYP2C9*3 is 6% in whites, 1% in African Americans, and 3% in Asians.19,20 People with CYP2C9*4 or CYP2C9*5 have a diminished capacity to clear warfarin; however, these variants occur so infrequently that their clinical relevance may be minimal.
Warfarin’s target, VKOR, is also polymorphic
Genetic variation in warfarin’s pharmacologic target, vitamin K 2,3-epoxide reductase (VKOR), also influences dose requirements. Warfarin decreases the synthesis of vitamin-K-dependent clotting factors by inhibiting VKOR. This inhibition depends on the patient’s C1 subunit gene, VKORC1. Patients with a guanine-to-adenine SNP 1,639 bases upstream of VKORC1 (−1639G>A) need lower warfarin doses—an average of 25% lower in those with the GA genotype (ie, one allele has guanine in the −1639 position and the other allele has adenine in that position) and 50% lower in those with the AA genotype compared with the wild-type genotype GG.21 This promoter SNP, positioned upstream (ie, before the gene-coding region), greatly influences VKORC1 expression.
A meta-analysis of 10 studies found that the VKORC1 polymorphism accounts for 25% of the interindividual variation in warfarin dose.18 In one study, the frequency of the AA genotype in a white population was 14%, AG 47%, and GG 39%; in a Chinese population the frequency of AA was 82%, AG 18%, and GG 0.35%.22
CYP4F2 and GGCX also affect warfarin’s dose requirements
Genetic variations in the enzymes CYP4F2 and gamma-glutamyl carboxylase (GGCX) also influence warfarin dose requirements. Although the data are limited and the effects are smaller than those of CYP2C9 and VKORC1, people with a SNP in CYP4F2 need 8% higher doses of warfarin, while those with a SNP in GGCX need 6% lower doses.23
CYP2C9 and VKORC1 testing is available
Currently, the clinical pharmacogenetic tests relevant for warfarin use are for CYP2C9 and VKORC1.10
The FDA has approved four warfarin pharmacogenetic test kits, but most third-party payers are reluctant to reimburse for testing because it is not currently considered a standard of care. Testing typically costs a few hundred dollars, but it should become less expensive as it becomes more commonplace. The current FDA-approved product label for warfarin does not recommend routine pharmacogenomic testing for determining initial or maintenance doses, but it does acknowledge that dose requirements are influenced by CYP2C9 and VKORC1 and states that genotype information, when available, can assist in selecting the starting dose.24
Clinical trials of warfarin pharmacogenomic testing are under way
Although genetic status can greatly influence an individual patient’s warfarin dosing requirement, routine prospective pharmacogenomic testing is not endorsed by the FDA or by other expert panels26 because there is currently insufficient evidence to recommend for or against it.
Several large prospective trials are under way. For example, the National Heart, Lung, and Blood Institute began a prospective trial in about 1,200 patients to evaluate the use of clinical plus genetic information to guide the initiation of warfarin therapy and to improve anticoagulation control for patients.27 The results, expected in September 2011, and those of other large prospective trials should provide adequate evidence for making recommendations about the clinical utility of routine pharmacogenetic testing for guiding warfarin therapy.
Several recent cost-utility and cost-effectiveness studies have attempted to quantify the value of pharmacogenomic testing for warfarin therapy,28–30 but their analyses are largely limited because the benefit (clinical utility) is yet to be sufficiently characterized.
The relevance of such analyses may soon be drastically diminished, as several non-vitamin-K-dependent blood thinners such as rivaroxaban (Xarelto), dabigatran (Pradaxa), and apixaban are poised to enter clinical practice.31
CLOPIDOGREL IS ACTIVATED BY CYP2C19
Clopidogrel, taken by about 40 million patients worldwide, is used to prevent atherothrombotic events and cardiac stent thrombosis when given along with aspirin.
Studies of clopidogrel pharmacogenomics
A recent genome-wide association study conducted in a cohort of 429 healthy Amish persons revealed a SNP in CYP2C19 to be associated with a diminished response to clopidogrel and to account for 12% of the variation in drug response.33 Traditional factors (the patient’s age, body-mass index, and cholesterol level) combined accounted for less than 10% of the variation.
Findings were similar in a subsequent investigation in 227 cardiac patients receiving clopidogrel: 21% of those with the variant had a cardiovascular ischemic event or died during a 1-year follow-up period compared with 10% of those without the variant (hazard ratio 2.42, P = .02).33
A 12-year prospective study investigating clopidogrel efficacy in 300 cardiac patients under the age of 45 used cardiovascular death, nonfatal myocardial infarction, and urgent coronary revascularization as end points. It concluded that the only independent predictor of these events was the patient’s CYP2C19 status.34
A study in 2,200 patients with recent myocardial infarction examined whether any of the known allelic variations that modulate clopidogrel’s absorption (ABCB1), metabolic activation (CYP3A4/5 and CYP2C19), or biologic activity (P2RY12 and ITGB3) was associated with a higher rate of the combined end point of all-cause mortality, nonfatal myocardial infarction, or stroke. None of the SNPs in CYP3A4/5, P2RY12, or ITGB3 that were evaluated was associated with a higher risk at 1 year. However, the allelic variations modulating clopidogrel’s absorption (ABCB1) and metabolism (CYP2C19) were associated with higher event rates. Patients with two variant ABCB1 alleles had a higher adjusted hazard ratio (95% confidence interval [CI] 1.2–2.47) than those with the wild-type allele. Patients who had one or two CYP2C19 loss-of-function alleles had a higher event rate than those with two wild-type alleles (95% CI 1.10–3.58 and 1.71–7.51, respectively).35
Conversely, researchers from the Population Health Research Institute found no association between poor-metabolizer status and treatment outcomes when CYP2C19 analysis was retrospectively added to the findings of two large clinical trials (combined N > 5,000). However, patients with acute coronary syndrome benefited more from clopidogrel treatment if they were ultra-rapid metabolizers (possessing the gain-of-function allele CYP2C19*17).36
Current status of clopidogrel testing: Uncertain
A current FDA boxed warning states that poor CYP2C19 metabolizers may not benefit from clopidogrel and recommends that prescribers consider alternative treatment for patients in this category.37 However, routine CYP2C19 testing is not recommended, and no firm recommendations have been established regarding dose adjustments for CYP2C19 status.
In 2010, the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association collectively pronounced the current evidence base insufficient for recommending routine pharmacogenomic testing.39
Needed are large-scale studies examining the cost-effectiveness and clinical utility of genotype-guided clopidogrel therapy compared with other therapy options such as prasugrel (Effient), an analogue not metabolized by CYP2C19. One such study, sponsored by Medco Health Solutions, plans to enroll 14,600 cardiac patients and has an estimated completion date in June 2011.40 The expectation that clopidogrel will be available in generic form in 2012 adds to the uncertainty regarding the cost-effectiveness of CYP2C19 testing for clopidogrel therapy.
STATINS: SLC01B1*5 INCREASES MYOPATHY RISK
Statins lower the concentration of low-density lipoprotein cholesterol (LDL-C), resulting in a relative-risk reduction of about 20% for each 1 mmol/L (39 mg/dL) decrement in LDL-C.41 They are one of the most commonly prescribed classes of drugs, but their side effects can limit their appeal: statin-induced myopathy occurs in about 1:1,000 to 1:10,000 patients and is difficult to predict.
SLC01B1. The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH), a genome-wide association study, recently found a SNP (SLCO1B1* 5) in the SLC01B1 gene to be associated with a higher risk of statin-induced myopathy in cardiac patients receiving simvastatin (Zocor) 40 or 80 mg daily.42 The SLC01B1 gene, located on chromosome 12, influences the extent of the drug’s hepatic uptake and its serum concentration. Only the SLC01B1*5 SNP emerged as a predictor of statin-induced myopathy across the entire genome.42
The authors believe the findings are likely to apply to other statins. The mechanisms leading to statin-induced myopathy and the impact of statin pharmacogenomics are still unclear.43
CYP3A4. Other genetic variants may play a vital role in determining response to statin therapy. Carriers of a newly identified CYP3A4 polymorphism (intron 6 SNP, rs35599367, C>T) required significantly lower statin doses (0.2–0.6 times less) for optimal lipid control. The analyses included atorvastatin (Lipitor), simvastatin, and lovastatin (Mevacor), and the association was robust (P = .019).44
Statin pharmacogenomic testing is not routinely recommended
Routine pharmacogenomic testing for statin therapy is not recommended. Additional studies are needed to determine the clinical utility and cost-effectiveness of pharmacogenomic testing (involving a combination of several polymorphisms) in various patient populations delineated by type of statin, dose, and concomitant use of other drugs.
TAMOXIFEN IS ACTIVATED BY CYP2D6
Tamoxifen is prescribed to prevent the recurrence of estrogen-receptor-positive breast cancer, to treat metastatic breast cancer, to prevent cancer in high-risk populations, and to treat ductal carcinoma in situ.
Tamoxifen is metabolized to form endoxifen, which has much higher potency and higher systemic levels than tamoxifen.45 Both CYP2D6 and CYP3A4/5 are required to produce endoxifen via two intermediates, but CYP2D6 catalyzes the critical step leading to metabolic activation.
The CYP2D6 gene is highly polymorphic, with more than 75 allelic variants identified. Extensive literature is available describing the influence of CYP2D6 polymorphisms on tamoxifen metabolism and therapy outcomes.46–52 Several CYP2D6 variants result in reduced or no enzyme activity, and people who have more than two normally functioning alleles have exaggerated enzyme activity (gene amplification).
Classification of CYP2D6 status
Several systems have been developed to categorize the phenotypic activity of CYP2D6 based on genotype.
A genetic basis for the observed diversity in the metabolism of cytochrome P450 substrates was recognized more than 30 years ago. People were categorized as either extensive or poor metabolizers, reflecting normal vs impaired ability to metabolize the CYP2D6 substrates sparteine and debrisoquine. Later work expanded this system to include categories for intermediate (between poor and extensive) and ultra-rapid (better than extensive) metabolizers.
The genetic basis for these categories includes homozygosity for dysfunctional variants (the poor-metabolizer group) or extra copies of normal functioning variants (the ultra-rapid-metabolizer group).
Newer systems have been described for characterizing the CYP2D6 activity phenotype whereby CYP2D6 variants are assigned activity scores.53–56 The various scoring systems have been reviewed by Kirchheiner.57
A recent version of the activity scoring system also takes into consideration the many drugs that inhibit CYP2D6, such as amiodarone (Cordarone) and fluoxetine (Prozac) that can reduce the action of tamoxifen if given with it (Table 4).58 For example, the tamoxifen exposure (as predicted by the CYP2D6-activity score) experienced by a CYP2D6 extensive metabolizer taking a CYP2D6-inhibiting drug may be similar to the exposure experienced by a CYP2D6 poor metabolizer receiving the same tamoxifen dose but not taking a CYP2D6-inhibiting drug.
Likewise, the activity score of a CYP2D6 intermediate metabolizer taking a CYP2D6-inducing drug may be similar to that of a CYP2D6 ultra-rapid metabolizer not taking a CYP2D6-inducing drug. Examples of CYP2D6 inducers are dexamethasone, rifampin, and hyperforin (St. John’s wort).
While the newer systems are reported to provide better correlations between genotype and phenotype scores, the older scoring systems and the categorical names are still widely used (eg, in the FDA-approved AmpliChip CYP450 test from Roche,59 which includes genotype data for CYP2D6 and CYP2C19).
No firm recommendations for CYP2D6 testing in tamoxifen users
The different genotypes and phenotypes vary in prevalence in different ethnic groups, and significantly different activity levels for endoxifen formation are observed. Tamoxifen lacks efficacy in those who are poor CYP2D6 metabolizers—ie, about 7% of the white population.
However, the FDA has not made firm recommendations about CYP2D6 testing for prescribing tamoxifen because the evidence of benefit, although suggestive, has been considered insufficient.
Clinicians should be aware that tamoxifen’s efficacy is greatly reduced by concomitant therapy with CYP2D6-inhibiting drugs (Table 4).
Other genes affecting tamoxifen: CYP3A4/5, SULT1A1, and UGT2B15
Some investigators propose that polymorphisms in additional genes encoding enzymes in the tamoxifen metabolic and elimination pathways (eg, CYP3A4/5, SULT1A1, and UGT2B15) also need to be considered to account adequately for interindividual variation in drug response.
For example, CYP3A4 and CYP3A5 are also polymorphic, and large interindividual variation exists in their enzyme activities. These enzymes have overlapping substrate specificities, represent the most abundant drug-metabolizing enzymes in the human liver, and are involved in the biotransformation of a broad range of endogenous substrates and most drugs.60
Clinical studies evaluating the impact of CYP3A4/5 polymorphisms have been inconsistent in their conclusions, which is generally attributed to the relatively low functional impact or the low prevalence of the SNPs evaluated. Many of the nearly 100 CYP3A4/5 polymorphisms identified have not yet been characterized regarding their functional impact on enzyme expression or activity. CYP-3A4/5 enzyme activity is highly variable between individuals and warrants further study of its role in outcomes of tamoxifen therapy. Ongoing and future prospective clinical trials evaluating CYP2D6, CYP3A4/5, and other relevant polymorphisms are necessary to define their clinical relevance before routine genetic testing for tamoxifen can be justified.
CODEINE IS ALSO ACTIVATED BY CYP2D6
Codeine also depends on the CYP2D6 gene, as it must be activated to its more potent opioid metabolites, including morphine. Poor CYP2D6 metabolizers do not benefit from codeine therapy.
The pharmacogenomics of codeine has become a hot topic, especially regarding breast-feeding mothers. The debate was ignited with the publication in 2006 of a case report of an infant’s death, apparently the result of metabolic polymorphisms.61 The evolution of this debate and the outcome of the case may be noteworthy to clinicians, as they illustrate the gravity of public and patient interest in pharmacogenomic testing. In this case, the breast-feeding mother had taken codeine regularly for about 14 days when her 13-day-old infant died from toxic levels of morphine. Unknown to her and the prescriber, both the mother and infant were ultra-rapid CYP2D6 metabolizers, resulting in a more rapid and extensive conversion of codeine to morphine.
A logical strategy for preventing similar deaths would be routine CYP2D6 genotyping when prescribing codeine to breast-feeding mothers. However, after several investigations examined the metabolic and excretion pathways of codeine in their entirety, the FDA did not recommend routine CYP2D6 testing when prescribing codeine to breastfeeding mothers because several other factors, including rare genetic variations of other enzymes, proved necessary for reaching the opioid toxicity leading to the infant’s death.62
PHARMACOGENOMICS OF PSYCHOTROPIC DRUGS
Pharmacogenomic testing has clinical utility for some psychotropic drugs.
HLA-B and carbamazepine
Considered a standard of care, HLA-B genotyping is appropriate before prescribing carbamazepine (Tegretol, Equetro) to patients in populations in which HLAB*1502 is likely to be present, such as Asians. Carriers of HLAB* 1502 are at higher risk of life-threatening skin reactions such as Stevens-Johnson syndrome.11
Several other pharmacogenomic applications for psychotropic medications have been suggested, but routine testing has not been recommended by the FDA or endorsed by any expert panel because sufficient clinical utility and cost-effectiveness have not been demonstrated. A brief summary of study findings and a few practical suggestions follow.
Polymorphisms in metabolizing enzymes have been investigated in patients receiving psychotropic drugs.
CYP2D6 and antidepressants
Many antidepressants show significant differences in plasma drug levels with CYP2D6 polymorphisms (in descending order of influence)55:
- Imipramine (Tofranil)
- Doxepin (Adapin, Silenor, Sinequan)
- Maprotiline (Deprilept, Ludiomil, Psymion)
- Trimipramine (Surmontil)
- Desipramine (Noraprim)
- Nortriptyline (Aventyl, Pamelor)
- Clomipramine (Anafranil)
- Paroxetine (Paxil)
- Venlafaxine (Effexor)
- Amitriptyline (Elavil)
- Mianserin
- Trazadone (Desyrel)
- Bupropion (Wellbutrin)
- Nefazodone (Serzone)
- Citalopram (Celexa)
- Sertraline (Zoloft).
CYP2D6 and antipsychotics
Several antipsychotics are also influenced by CYP2D6 polymorphisms (also in descending order of influence)55:
- Perphenazine (Trilafon)
- Thioridazine (Mellaril)
- Olanzapine (Zyprexa)
- Zuclopenthixol (Cisordinol, Clopixol, Acuphase)
- Aripiprazole (Abilify)
- Flupentixol (Depixol, Fluanxol)
- Haloperidol (Haldol)
- Perazine (Taxilan)
- Risperidone (Risperdal)
- Pimozide (Orap).
CYP2C19 and antidepressants
CYP2C19 polymorphisms are likewise associated with differences in drug metabolism for many antidepressants, such as (in descending order of CYP2C19-mediated influence)55:
- Trimipramine
- Doxepin
- Amitriptyline
- Imipramine
- Citalopram (Celexa)
- Clomipramine
- Moclobemide (Aurorix, Manerix)
- Sertraline
- Fluvoxamine (Luvox).
Clinical relevance of CYP2D6 and CYP2C19
Several studies have demonstrated that poor and intermediate CYP2D6 metabolizers have a higher incidence of adverse effects when taking CYP2D6-dependent antidepressants63–68; however, an almost equal number of studies did not find statistically significant associations.69–72 Likewise, several studies have found an association between ultra-rapid CYP2D6 metabolizer status and diminished response to antidepressants,65,73,74 but no association was found in a larger retrospective study.75
Routine CYP2D6 and CYP2C19 screening is not recommended when prescribing psychotropic drugs. However, reviews of the pharmacokinetic data have suggested a few practical applications when genetic status is already known. In general, clinicians can consider reducing the dose of tricyclic antidepressants by about 50% when prescribing to CYP2D6-poor-metabolizers.55,76–78
Genes that affect serotonin metabolism
Several genes in the serotonin pathway have been investigated to determine whether they influence patients’ susceptibility to depression and adverse effects and response to psychotropic medications.
SLC6A4. Polymorphisms in the promoter region of the serotonin transporter gene SLC6A4 appear to influence the treatment response and side-effect profiles of selective serotonin reuptake inhibitors (SSRIs). Carriers of the SLC6A4 5-HTTLPR L alleles have fewer side effects79 and better response to SSRI treatment, and carriers of the S allele have a higher incidence of antidepressant-induced mania80 and poorer response to SSRI treatment.81
5-HT. Polymorphisms in serotonin receptors (2A and 2C subtypes) appear to influence SSRI response and side effects. Carriers of 5-HT 2A C alleles had more severe adverse effects from paroxetine,71 but another 5-HT 2A polymorphism common to Asians is associated with better response to antidepressant therapy.82 A 5-HT 2C polymorphism was associated with a lower incidence of antipsychotic-induced weight gain.83
Although the understanding of these relationships is incomplete and routine pharmacogenomic testing is not currently recommended, reviews of the pharmacodynamic data have suggested a few practical applications when a patient’s genetic status is already known. One should consider:
- Selecting treatments other than SSRIs for depressed patients known to possess the SLC6A4 variant
- Selecting citalopram for depressed patients known to carry the 5-HT 2A polymorphism
- Avoiding treatment with antipsychotic drugs for patients known to possess the 5-HT 2C polymorphism.
THE FUTURE OF PHARMACOGENOMIC TESTING
The examples discussed in this article provide some insight about how pharmacogenomic testing is maturing and slowly being integrated into the practice of medicine. They also illustrate the complexity of the multiple stages of research that pharmacogenomic applications must go through in order to be adopted as standard practice.
In the future, pharmacogenomic data will continue to accumulate, and the clinical utility of many other pharmacogenomic tests may be uncovered. The FDA provides information on emerging pharmacogenomic tests at its Web site, www.fda.gov.11 Its up-to-date “Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels” includes boxed warnings, recommendations, research outcomes, and relevant population genetics.
If the FDA continues its current policy, prospective randomized trials that show improvement in patient outcomes will remain the gold standard for determining the clinical significance of a pharmacogenomic test. Furthermore, cost-benefit analyses are likely to continue dictating policy regarding pharmacogenomic testing, and cost-benefit profiles should improve as technology advances and as information gathered from a single test becomes applicable to multiple medications and clinical scenarios.
In the meantime, physicians should become familiar with the terms used in medical genetics and pharmacogenomics and begin to understand genetic contributions to the outcomes of drug therapy. For example, understanding the consequences of metabolizer status and the frequency of variants in a given population can be tremendously helpful when advising our patients about anticipating potential problems when taking specific medications and about making informed decisions about pharmacogenomic testing.
This exchange of information alone may go a long way in improving therapy outcomes even when prospective pharmacogenomic testing is not routinely performed. Furthermore, an increasing number of patients will already have genotyping information available when they come to us, and clinicians need to be aware of the many pharmacogenomic applications recommended by the FDA when genetic status is known.10
- King HC, Sinha AA. Gene expression profile analysis by DNA micro-arrays: promise and pitfalls. JAMA 2001; 286:2280–2288.
- Nuckols TK, Paddock SM, Bower AG, et al. Costs of intravenous adverse drug events in academic and nonacademic intensive care units. Med Care 2008; 46:17–24.
- Gurwitz JH, Field TS, Judge J, et al. The incidence of adverse events in two long-term care facilities. Am J Med 2005; 118:251–258.
- Vargas E, Terleira A, Hernando F, et al. Effects of adverse drug reactions on length of stay in surgical intensive care units. Crit Care Med 2003; 31:694–698.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998; 279:1200–1205.
- Field TS, Gilman BH, Subramanian S, Fuller JC, Bates DW, Gurwitz JH. The costs associated with adverse drug events in older adults in the ambulatory setting. Med Care 2005; 43:1171–1176.
- Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA 1997; 277:301–306.
- Ernst FR, Grizzle AJ. Drug-related morbidity and mortality: updating the cost-of-illness model. J Am Pharm Assoc (Wash) 2001; 41:192–199.
- Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002; 3:229–243.
- Sadee W. Measuring cis-acting regulatory variants genome-wide: new insights into expression genetics and disease susceptibility. Genome Med 2009; 1:116.
- US Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labels. http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm. Accessed 1/18/2011.
- Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med 2007; 147:755–765.
- Elias DJ, Topol EJ. Warfarin pharmacogenomics: a big step forward for individualized medicine: enlightened dosing of warfarin. Eur J Hum Genet 2008; 16:532–534.
- Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study). J Am Coll Cardiol 2010; 55:2804–2812.
- Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 1994; 4:39–42.
- Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002; 287:1690–1698.
- Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med 2005; 7:97–104.
- Au N, Rettie AE. Pharmacogenomics of 4-hydroxycoumarin anticoagulants. Drug Metab Rev 2008; 40:355–375.
- García-Martín E, Martínez C, Ladero JM, Agúndez JA. Interethnic and intraethnic variability of CYP2C8 and CYP2C9 polymorphisms in healthy individuals. Mol Diagn Ther 2006; 10:29–40.
- Sullivan-Klose TH, Ghanayem BI, Bell DA, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics 1996; 6:341–349.
- Wen MS, Lee M, Chen JJ, et al. Prospective study of warfarin dosage requirements based on CYP2C9 and VKORC1 genotypes. Clin Pharmacol Ther 2008; 84:83–89.
- Larramendy-Gozalo C, Yang JQ, Verstuyft C, et al. Genetic polymorphism of vitamin K epoxide reductase (VKORC1) 1173C>T in a Chinese and a Caucasian population. Basic Clin Pharmacol Toxicol 2006; 98:611–613.
- Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood 2008; 111:4106–4112.
- Bristol-Myers Squibb. Coumadin (warfarin sodium) Prescribing Information. January 2010.
- Barnes-Jewish Hospital at Washington University Medical Center. Warfarin dosing. http://warfarindosing.orgAccessed 1/20/2011.
- Flockhart DA, O’Kane D, Williams MS, et al. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med 2008; 10:139–150.
- National Institutes of Health. ClinicalTrials.gov. http://clinicaltrials.gov. Accessed January 20, 2011.
- Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med 2009; 150:73–83.
- Meckley LM, Gudgeon JM, Anderson JL, Williams MS, Veenstra DL. A policy model to evaluate the benefits, risks, and costs of warfarin pharmacogenomic testing. Pharmacoeconomics 2010; 28:61–74.
- Patrick AR, Avron J, Choudhry NK. Cost-effectiveness of genotype-guided warfarin dosing for patients with atrial fibrillation. Circ Cardiovasc Qual Outcomes 2009; 2:429–436.
- Haas S. New oral Xa and IIa inhibitors: updates on clinical trial results. J Thromb Thrombolysis 2008; 25:52–60.
- Bhatt DL. Tailoring antiplatelet therapy based on pharmacogenomics: how well do the data fit? JAMA 2009; 302:896–897.
- Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009; 302:849–857.
- Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 2009; 373:309–317.
- Simon T, Verstuyft C, Mary-Krause M, et al; French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009; 360:363–375.
- Paré G, Mehta SR, Yusuf S, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med 2010; 363:1704–1714.
- Bristol-Myers Squibb/Sanofi Pharmaceutical Partnership. Plavix (clopidogrel bisulfate) prescribing information. August 2010.
- P450 Drug Interaction Table. Indiana University School of Medicine. http://medicine.iupui.edu/clinpharm/ddis/table.asp. Accessed 1/21/2011.
- Society for Cardiovascular Angiography and Interventions; Holmes DR, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation 2010; 122:537–557.
- National Institutes of Health. Genotype Guided Comparison of Clopidogrel and Prasugrel Outcomes Study. http://clinicaltrialsfeeds.org/clinical-trials/show/NCT00995514. Accessed 1/20/2011.
- Amarenco P, Labreuche J. Lipid management in the prevention of stroke: Review and updated meta-analysis of statins for stroke prevention. Lancet Neurol 2009; 8:453–463.
- SEARCH Collaborative Group, Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med 2008; 359:789–799.
- Romaine SP, Bailey KM, Hall AS, Balmforth AJ. The influence of SLC01B1 (OATP1B1) gene polymorphisms on response to statin therapy. Pharmacogenomics J 2010; 10:1–11.
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 2010; Apr 13 [Epub ahead of print].
- Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 2005; 23:9312–9318.
- Beverage JN, Sissung TM, Sion AM, Danesi R, Figg WD. CYP2D6 polymorphisms and the impact on tamoxifen therapy. J Pharm Sci 2007; 96:2224–2231.
- Flockhart D. CYP2D6 genotyping and the pharmacogenetics of tamoxifen. Clin Adv Hematol Oncol 2008; 6:493–494.
- Goetz MP, Kamal A, Ames MM. Tamoxifen pharmacogenomics: the role of CYP2D6 as a predictor of drug response. Clin Pharmacol Ther 2008; 83:160–166.
- Stearns V, Rae JM. Pharmacogenetics and breast cancer endocrine therapy: CYP2D6 as a predictive factor for tamoxifen metabolism and drug response? Expert Rev Mol Med 2008; 10:e34.
- Dezentjé VO, Guchelaar HJ, Nortier JW, van del Velde CJ, Gelderblom H. Clinical implications of CYP2D6 genotyping in tamoxifen treatment for breast cancer. Clin Cancer Res 2009; 15:15–21.
- Higgins MJ, Rae JM, Flockhart DA, Hayes DF, Stearns V. Pharmacogenetics of tamoxifen: who should undergo CYP2D6 genetic testing? J Natl Compr Canc Netw 2009; 7:203–213.
- Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer 2009; 9:576–586.
- Steimer W, Zöpf K, von Amelunxen S, et al. Allele-specific change of concentration and functional gene dose for the prediction of steady-state serum concentrations of amitriptyline and nortriptyline in CYP2C19 and CYP2D6 extensive and intermediate metabolizers. Clinical Cancer 2004; 50:1623–1633.
- Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharm Ther 2008; 83:234–242.
- Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry 2004; 9:442–473.
- Kirchheiner J, Schmidt H, Tzvetkov M, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J 2007; 7:257–265.
- Kirchheiner J. CYP2D6 phenotype prediction from genotype: which system is the best? Clin Pharmacol Ther 2008; 83:225–227.
- Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol 2010; 50:450–458.
- Hoffmann-La Roche Ltd. AmpliChip CYP450 Test. http://www.roche.com/assays/Pages/AmpliChipCYP450Test.aspx. Accessed 1/21/2011.
- Anzenbacher P, Anzenbacherová E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci 2001; 58:737–747.
- Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet 2006; 368:704.
- Willmann S, Edginton AN, Coboeken K, Ahr G, Lippert J. Risk to the breast-fed neonate from codeine treatment to the mother: a quantitative mechanistic modeling study. Clin Pharm Ther 2009; 86:634–643.
- Grzesiak M, Beszłej A, Lebioda A, Jonkisz A, Dobosz T, Kienja A. [Retrospective assessment of the antidepressants tolerance in the group of patients with diagnosis of depression and different CYP2D6 genotype.] [In Polish] Psychiatr Pol 2003; 37:433–444.
- Laika B, Leucht S, Heres S, Steimer W. Intermediate metabolizer: increased side effects in psychoactive drug therapy. The key to cost-effectiveness of pretreatment CYP2D6 screening? Pharmacogenomics J 2009; 9:395–403.
- Rau T, Wohlleben G, Wuttke H, et al. CYP2D6 genotype: Impact on adverse effects and nonresponse during treatment with antidepressants—a pilot study. Clin Pharm Ther 2004; 75:386–393.
- McAlpine DE, O’Kane DJ, Black JL, Mrazek DA. Cytochrome P450 2D6 genotype variation and venlafaxine dosage. Mayo Clin Proc 2007; 82:1065–1068.
- Chen S, Chou WH, Blouin RA, et al. The cytochrome P450 2D6 (CYP2D6) enzyme polymorphism: screening costs and influence on clinical outcomes in psychiatry. Clin Pharmacol Ther 1996; 60:522–534.
- Shams ME, Arneth B, Hiemke C, et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther 2006; 31:493–502.
- Whyte EM, Romkes M, Mulsant BH, et al. CYP2D6 genotype and venlafaxine-XR concentrations in depressed elderly. Int J Geriatr Psychiatry 2006; 21:542–549.
- Roberts RL, Mulder RT, Joyce PR, Luty SE, Kennedy MA. No evidence of increased adverse drug reactions in cytochrome P450 CYP2D6 poor metabolizers treated with fluoxetine or nortriptyline. Hum Psychopharmacol 2004; 19:17–23.
- Murphy GM, Kremer C, Rodrigues HE, Schatzberg AF. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry 2003; 160:1830–1835.
- Gillman PK. Re: no evidence of increased adverse drug reactions in cytochrome P450 CYP2D6 poor metabolizers treated with fluoxetine or nortriptyline. Hum Psychopharmacol 2005; 20:61–62.
- Gex-Fabry M, Eap CB, Oneda B, et al. CYP2D6 and ABCB1 genetic variability: influence on paroxetine plasma level and therapeutic response. Ther Drug Monit 2008; 30:474–482.
- Kawanishi C, Lundgren S, Agren H, Bertilsson L. Increased incidence of CYP2D6 gene duplication in patients with persistent mood disorders: ultrarapid metabolism of antidepressants as a cause of nonresponse. A pilot study. Eur J Clin Pharmacol 2004; 59:803–807.
- Serretti A, Calati R, Massat I, et al. Cytochrome P450 CYP1A2, CYP2C9, CYP2C19 and CYP2D6 genes are not associated with response and remission in a sample of depressive patients. Int Clin Psychopharmacol 2009; 24:250–256.
- de Leon J, Armstrong SC, Cozza KL. Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics 2006; 47:75–85.
- de Leon J, Susce MT, Johnson M, et al. DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr 2009; 14:19–34.
- Thuerauf N, Lunkenheimer J. The impact of the CYP2D6-polymorphism on dose recommendations for current antidepressants. Eur Arch Psychiatry Clin Neurosci 2006; 256:287–293.
- Horstmann S, Binder EB. Pharmacogenomics of antidepressant drugs. Pharmacol Ther 2009; 124:57–73.
- Ferreira Ade A, Neves FS, da Rocha FF, et al. The role of 5-HTTLPR polymorphism in antidepressant-associated mania in bipolar disorder. J Affect Disord 2009; 112:267–272.
- Serretti A, Kato M, De Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry 2007; 12:247–257.
- Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry 2010; 15:473–500.
- Reynolds GP, Zhang Z, Zhang X. Polymorphism of the promoter region of the serotonin 5-HT(2C) receptor gene and clozapine-induced weight gain. Am J Psychiatry 2003; 160:677–679.
In many patients, certain drugs do not work as well as expected, whereas in other patients they cause toxic effects, even at lower doses. For some patients, the reason may be genetic.
Sizeable minorities of the population carry genetic variants—polymorphisms— that affect their response to various drugs. Thanks to genetic research, our understanding of the variability of drug response has advanced markedly in the last decade. Many relevant polymorphisms have been identified, and tests for some of them are available.
Armed with the knowledge of their patients’ genetic status, physicians could predict their response to certain drugs, leading to better efficacy, fewer adverse drug reactions, and a better cost-benefit ratio.
The possible impact is substantial, since more than half of the drugs most commonly involved in adverse drug reactions are metabolized by polymorphic enzymes.1 Adverse drug reactions remain a significant detriment to public health, having a substantial impact on rates of morbidity and death and on healthcare costs.2–8 In the United States, adverse drug reactions are a leading cause of death in hospitalized patients4 and are annually responsible for hundreds of thousands of deaths and hundreds of billions of dollars in added costs.2,4,6–8
In the meantime, physicians can educate their patients and promote efforts to incorporate genomic information into standard clinical decision-making.
This article offers an overview of pharmacogenomic testing, discussing implications and limitations of a few validated tests. Specifically, we will discuss testing that is relevant when using warfarin (Coumadin), clopidogrel (Plavix), statins, tamoxifen (Nolvadex), codeine, and psychotropic medications, as well as the future role of pharmacogenomic testing in medicine.
WHAT IS PHARMACOGENOMICS?
Pharmacogenomics is the study of how genetic factors relate to interindividual variability of drug response.
Many clinicians may not be familiar with the background and terminology used in the pharmacogenomic literature. Below, a brief review of the terminology is followed by a schematic describing the various stages of research involved in pharmacogenomics and the advancement of a test into standard practice.
The review and schematic may be helpful for evaluating the clinical significance of pharmacogenomics-related articles.
From genotype to phenotype
Genotype refers to the coding sequence of DNA base pairs for a particular gene, and phenotype (eg, disease or drug response) refers to a trait resulting from the protein product encoded by the gene. The name of a gene often refers to its protein product and is italicized (eg, the CYP3A4 gene encodes for the CYP3A4 enzyme).
Two alleles per autosomal gene (one paternal and one maternal) form the genotype. Heterozygotes possess two different alleles, and homozygotes possess two of the same alleles. The most common allele in a population is referred to as the wild type, and allele frequencies can vary greatly in different populations.9
Most sequence variations are single nucleotide polymorphisms (SNPs, pronounced “snips”), a single DNA base pair substitution that may result in a different gene product. SNPs can be classified as structural RNA polymorphisms (srSNPs), regulatory polymorphisms (rSNPs), or polymorphisms in coding regions (cSNPs)10: srSNPs alter mRNA processing and translation, rSNPs alter transcription, and cSNPs alter protein sequence and function.
Recently, genetic associations with a phenotype have been done on a large scale, with millions of SNPs measured in each of many subjects. This approach, called a genomewide association study or GWAS, has revealed countless candidate genes for clinical traits, but only a few have resulted in a practical clinical application. SNPs may by themselves exert a pharmacokinetic effect (ie, how the body processes the drug), a pharmacodynamic effect (ie, how the drug affects the body), or both, or they may act in concert with other genetic factors. Pharmacodynamic effects can result from a pharmacokinetic effect or can result from variations in a pharmacologic target.
Establishing a genotype-phenotype association can involve clinical studies, animal transgenic studies, or molecular and cellular functional assays.
Clinical applications are emerging
WARFARIN: IMPORTANCE OF CYP2C9, VKORC1
Warfarin is used for the long-term treatment and prevention of thromboembolic events.
This drug has a narrow therapeutic window and shows substantial interpatient dose variability. The start of warfarin therapy is associated with one of the highest rates of adverse events and emergency room visits of any single drug.12 More than 2 million patients start warfarin each year in the United States alone,13 and about 20% of them are hospitalized within the first 6 months because of bleeding due to overanticoagulation.14
The findings from a recent study suggest that pharmacogenomic testing may eventually allow more patients to safely benefit from warfarin therapy. In this large, nationwide, prospective study, hospitalization rates were 30% lower when pharmacogenomic testing was used.14 However, no reduction was seen in the time needed to reach the target international normalized ratio (INR) or in the need for INR checks at 6 months. Furthermore, this study used historical control data, and some or all of the reduction in hospitalization rates may be attributed to more frequent INR checks in the patients who underwent testing than in the historical control group.
A relationship between warfarin dose requirements and the genetic status of CYP2C9, which encodes a major drug-metabolizing enzyme, has been demonstrated in retrospective and prospective studies.15–17
S-warfarin is metabolized by CYP2C9, which is polymorphic
Warfarin contains equal amounts of two isomers, designated S and R. S-warfarin, which is more potent, is metabolized principally by CYP2C9, while R-warfarin is metabolized by CYP1A2, CYP2C19, and CYP3A4.
People who possess two copies of the wild type CYP2C9 gene CYP2C9*1 metabolize warfarin very well and so are called “extensive warfarin metabolizers.” Carriers of the allelic variants CYP2C9*2 and CYP2C9*3 (which have point mutations in exons 3 and 7 of CYP2C9, respectively), have less capacity. Compared with those who are homozygous for the wild-type gene, homozygous carriers of CYP2C9* 3 clear S-warfarin at a rate that is 90% lower, and those with the CYP2C9*1/*3, CYP2C9* 1/*2, CYP2C9*2/*2, or CYP2C9*2/*3 genotypes clear it at a rate 50% to 75% lower. A meta-analysis of 12 studies found that the CYP2C9 genotype accounted for 12% of the interindividual variability of warfarin dose requirements.18
About 8% of whites carry at least one copy of CYP2C9*2, as do 1% of African Americans; the allele is rare in Asian populations. The frequency of CYP2C9*3 is 6% in whites, 1% in African Americans, and 3% in Asians.19,20 People with CYP2C9*4 or CYP2C9*5 have a diminished capacity to clear warfarin; however, these variants occur so infrequently that their clinical relevance may be minimal.
Warfarin’s target, VKOR, is also polymorphic
Genetic variation in warfarin’s pharmacologic target, vitamin K 2,3-epoxide reductase (VKOR), also influences dose requirements. Warfarin decreases the synthesis of vitamin-K-dependent clotting factors by inhibiting VKOR. This inhibition depends on the patient’s C1 subunit gene, VKORC1. Patients with a guanine-to-adenine SNP 1,639 bases upstream of VKORC1 (−1639G>A) need lower warfarin doses—an average of 25% lower in those with the GA genotype (ie, one allele has guanine in the −1639 position and the other allele has adenine in that position) and 50% lower in those with the AA genotype compared with the wild-type genotype GG.21 This promoter SNP, positioned upstream (ie, before the gene-coding region), greatly influences VKORC1 expression.
A meta-analysis of 10 studies found that the VKORC1 polymorphism accounts for 25% of the interindividual variation in warfarin dose.18 In one study, the frequency of the AA genotype in a white population was 14%, AG 47%, and GG 39%; in a Chinese population the frequency of AA was 82%, AG 18%, and GG 0.35%.22
CYP4F2 and GGCX also affect warfarin’s dose requirements
Genetic variations in the enzymes CYP4F2 and gamma-glutamyl carboxylase (GGCX) also influence warfarin dose requirements. Although the data are limited and the effects are smaller than those of CYP2C9 and VKORC1, people with a SNP in CYP4F2 need 8% higher doses of warfarin, while those with a SNP in GGCX need 6% lower doses.23
CYP2C9 and VKORC1 testing is available
Currently, the clinical pharmacogenetic tests relevant for warfarin use are for CYP2C9 and VKORC1.10
The FDA has approved four warfarin pharmacogenetic test kits, but most third-party payers are reluctant to reimburse for testing because it is not currently considered a standard of care. Testing typically costs a few hundred dollars, but it should become less expensive as it becomes more commonplace. The current FDA-approved product label for warfarin does not recommend routine pharmacogenomic testing for determining initial or maintenance doses, but it does acknowledge that dose requirements are influenced by CYP2C9 and VKORC1 and states that genotype information, when available, can assist in selecting the starting dose.24
Clinical trials of warfarin pharmacogenomic testing are under way
Although genetic status can greatly influence an individual patient’s warfarin dosing requirement, routine prospective pharmacogenomic testing is not endorsed by the FDA or by other expert panels26 because there is currently insufficient evidence to recommend for or against it.
Several large prospective trials are under way. For example, the National Heart, Lung, and Blood Institute began a prospective trial in about 1,200 patients to evaluate the use of clinical plus genetic information to guide the initiation of warfarin therapy and to improve anticoagulation control for patients.27 The results, expected in September 2011, and those of other large prospective trials should provide adequate evidence for making recommendations about the clinical utility of routine pharmacogenetic testing for guiding warfarin therapy.
Several recent cost-utility and cost-effectiveness studies have attempted to quantify the value of pharmacogenomic testing for warfarin therapy,28–30 but their analyses are largely limited because the benefit (clinical utility) is yet to be sufficiently characterized.
The relevance of such analyses may soon be drastically diminished, as several non-vitamin-K-dependent blood thinners such as rivaroxaban (Xarelto), dabigatran (Pradaxa), and apixaban are poised to enter clinical practice.31
CLOPIDOGREL IS ACTIVATED BY CYP2C19
Clopidogrel, taken by about 40 million patients worldwide, is used to prevent atherothrombotic events and cardiac stent thrombosis when given along with aspirin.
Studies of clopidogrel pharmacogenomics
A recent genome-wide association study conducted in a cohort of 429 healthy Amish persons revealed a SNP in CYP2C19 to be associated with a diminished response to clopidogrel and to account for 12% of the variation in drug response.33 Traditional factors (the patient’s age, body-mass index, and cholesterol level) combined accounted for less than 10% of the variation.
Findings were similar in a subsequent investigation in 227 cardiac patients receiving clopidogrel: 21% of those with the variant had a cardiovascular ischemic event or died during a 1-year follow-up period compared with 10% of those without the variant (hazard ratio 2.42, P = .02).33
A 12-year prospective study investigating clopidogrel efficacy in 300 cardiac patients under the age of 45 used cardiovascular death, nonfatal myocardial infarction, and urgent coronary revascularization as end points. It concluded that the only independent predictor of these events was the patient’s CYP2C19 status.34
A study in 2,200 patients with recent myocardial infarction examined whether any of the known allelic variations that modulate clopidogrel’s absorption (ABCB1), metabolic activation (CYP3A4/5 and CYP2C19), or biologic activity (P2RY12 and ITGB3) was associated with a higher rate of the combined end point of all-cause mortality, nonfatal myocardial infarction, or stroke. None of the SNPs in CYP3A4/5, P2RY12, or ITGB3 that were evaluated was associated with a higher risk at 1 year. However, the allelic variations modulating clopidogrel’s absorption (ABCB1) and metabolism (CYP2C19) were associated with higher event rates. Patients with two variant ABCB1 alleles had a higher adjusted hazard ratio (95% confidence interval [CI] 1.2–2.47) than those with the wild-type allele. Patients who had one or two CYP2C19 loss-of-function alleles had a higher event rate than those with two wild-type alleles (95% CI 1.10–3.58 and 1.71–7.51, respectively).35
Conversely, researchers from the Population Health Research Institute found no association between poor-metabolizer status and treatment outcomes when CYP2C19 analysis was retrospectively added to the findings of two large clinical trials (combined N > 5,000). However, patients with acute coronary syndrome benefited more from clopidogrel treatment if they were ultra-rapid metabolizers (possessing the gain-of-function allele CYP2C19*17).36
Current status of clopidogrel testing: Uncertain
A current FDA boxed warning states that poor CYP2C19 metabolizers may not benefit from clopidogrel and recommends that prescribers consider alternative treatment for patients in this category.37 However, routine CYP2C19 testing is not recommended, and no firm recommendations have been established regarding dose adjustments for CYP2C19 status.
In 2010, the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association collectively pronounced the current evidence base insufficient for recommending routine pharmacogenomic testing.39
Needed are large-scale studies examining the cost-effectiveness and clinical utility of genotype-guided clopidogrel therapy compared with other therapy options such as prasugrel (Effient), an analogue not metabolized by CYP2C19. One such study, sponsored by Medco Health Solutions, plans to enroll 14,600 cardiac patients and has an estimated completion date in June 2011.40 The expectation that clopidogrel will be available in generic form in 2012 adds to the uncertainty regarding the cost-effectiveness of CYP2C19 testing for clopidogrel therapy.
STATINS: SLC01B1*5 INCREASES MYOPATHY RISK
Statins lower the concentration of low-density lipoprotein cholesterol (LDL-C), resulting in a relative-risk reduction of about 20% for each 1 mmol/L (39 mg/dL) decrement in LDL-C.41 They are one of the most commonly prescribed classes of drugs, but their side effects can limit their appeal: statin-induced myopathy occurs in about 1:1,000 to 1:10,000 patients and is difficult to predict.
SLC01B1. The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH), a genome-wide association study, recently found a SNP (SLCO1B1* 5) in the SLC01B1 gene to be associated with a higher risk of statin-induced myopathy in cardiac patients receiving simvastatin (Zocor) 40 or 80 mg daily.42 The SLC01B1 gene, located on chromosome 12, influences the extent of the drug’s hepatic uptake and its serum concentration. Only the SLC01B1*5 SNP emerged as a predictor of statin-induced myopathy across the entire genome.42
The authors believe the findings are likely to apply to other statins. The mechanisms leading to statin-induced myopathy and the impact of statin pharmacogenomics are still unclear.43
CYP3A4. Other genetic variants may play a vital role in determining response to statin therapy. Carriers of a newly identified CYP3A4 polymorphism (intron 6 SNP, rs35599367, C>T) required significantly lower statin doses (0.2–0.6 times less) for optimal lipid control. The analyses included atorvastatin (Lipitor), simvastatin, and lovastatin (Mevacor), and the association was robust (P = .019).44
Statin pharmacogenomic testing is not routinely recommended
Routine pharmacogenomic testing for statin therapy is not recommended. Additional studies are needed to determine the clinical utility and cost-effectiveness of pharmacogenomic testing (involving a combination of several polymorphisms) in various patient populations delineated by type of statin, dose, and concomitant use of other drugs.
TAMOXIFEN IS ACTIVATED BY CYP2D6
Tamoxifen is prescribed to prevent the recurrence of estrogen-receptor-positive breast cancer, to treat metastatic breast cancer, to prevent cancer in high-risk populations, and to treat ductal carcinoma in situ.
Tamoxifen is metabolized to form endoxifen, which has much higher potency and higher systemic levels than tamoxifen.45 Both CYP2D6 and CYP3A4/5 are required to produce endoxifen via two intermediates, but CYP2D6 catalyzes the critical step leading to metabolic activation.
The CYP2D6 gene is highly polymorphic, with more than 75 allelic variants identified. Extensive literature is available describing the influence of CYP2D6 polymorphisms on tamoxifen metabolism and therapy outcomes.46–52 Several CYP2D6 variants result in reduced or no enzyme activity, and people who have more than two normally functioning alleles have exaggerated enzyme activity (gene amplification).
Classification of CYP2D6 status
Several systems have been developed to categorize the phenotypic activity of CYP2D6 based on genotype.
A genetic basis for the observed diversity in the metabolism of cytochrome P450 substrates was recognized more than 30 years ago. People were categorized as either extensive or poor metabolizers, reflecting normal vs impaired ability to metabolize the CYP2D6 substrates sparteine and debrisoquine. Later work expanded this system to include categories for intermediate (between poor and extensive) and ultra-rapid (better than extensive) metabolizers.
The genetic basis for these categories includes homozygosity for dysfunctional variants (the poor-metabolizer group) or extra copies of normal functioning variants (the ultra-rapid-metabolizer group).
Newer systems have been described for characterizing the CYP2D6 activity phenotype whereby CYP2D6 variants are assigned activity scores.53–56 The various scoring systems have been reviewed by Kirchheiner.57
A recent version of the activity scoring system also takes into consideration the many drugs that inhibit CYP2D6, such as amiodarone (Cordarone) and fluoxetine (Prozac) that can reduce the action of tamoxifen if given with it (Table 4).58 For example, the tamoxifen exposure (as predicted by the CYP2D6-activity score) experienced by a CYP2D6 extensive metabolizer taking a CYP2D6-inhibiting drug may be similar to the exposure experienced by a CYP2D6 poor metabolizer receiving the same tamoxifen dose but not taking a CYP2D6-inhibiting drug.
Likewise, the activity score of a CYP2D6 intermediate metabolizer taking a CYP2D6-inducing drug may be similar to that of a CYP2D6 ultra-rapid metabolizer not taking a CYP2D6-inducing drug. Examples of CYP2D6 inducers are dexamethasone, rifampin, and hyperforin (St. John’s wort).
While the newer systems are reported to provide better correlations between genotype and phenotype scores, the older scoring systems and the categorical names are still widely used (eg, in the FDA-approved AmpliChip CYP450 test from Roche,59 which includes genotype data for CYP2D6 and CYP2C19).
No firm recommendations for CYP2D6 testing in tamoxifen users
The different genotypes and phenotypes vary in prevalence in different ethnic groups, and significantly different activity levels for endoxifen formation are observed. Tamoxifen lacks efficacy in those who are poor CYP2D6 metabolizers—ie, about 7% of the white population.
However, the FDA has not made firm recommendations about CYP2D6 testing for prescribing tamoxifen because the evidence of benefit, although suggestive, has been considered insufficient.
Clinicians should be aware that tamoxifen’s efficacy is greatly reduced by concomitant therapy with CYP2D6-inhibiting drugs (Table 4).
Other genes affecting tamoxifen: CYP3A4/5, SULT1A1, and UGT2B15
Some investigators propose that polymorphisms in additional genes encoding enzymes in the tamoxifen metabolic and elimination pathways (eg, CYP3A4/5, SULT1A1, and UGT2B15) also need to be considered to account adequately for interindividual variation in drug response.
For example, CYP3A4 and CYP3A5 are also polymorphic, and large interindividual variation exists in their enzyme activities. These enzymes have overlapping substrate specificities, represent the most abundant drug-metabolizing enzymes in the human liver, and are involved in the biotransformation of a broad range of endogenous substrates and most drugs.60
Clinical studies evaluating the impact of CYP3A4/5 polymorphisms have been inconsistent in their conclusions, which is generally attributed to the relatively low functional impact or the low prevalence of the SNPs evaluated. Many of the nearly 100 CYP3A4/5 polymorphisms identified have not yet been characterized regarding their functional impact on enzyme expression or activity. CYP-3A4/5 enzyme activity is highly variable between individuals and warrants further study of its role in outcomes of tamoxifen therapy. Ongoing and future prospective clinical trials evaluating CYP2D6, CYP3A4/5, and other relevant polymorphisms are necessary to define their clinical relevance before routine genetic testing for tamoxifen can be justified.
CODEINE IS ALSO ACTIVATED BY CYP2D6
Codeine also depends on the CYP2D6 gene, as it must be activated to its more potent opioid metabolites, including morphine. Poor CYP2D6 metabolizers do not benefit from codeine therapy.
The pharmacogenomics of codeine has become a hot topic, especially regarding breast-feeding mothers. The debate was ignited with the publication in 2006 of a case report of an infant’s death, apparently the result of metabolic polymorphisms.61 The evolution of this debate and the outcome of the case may be noteworthy to clinicians, as they illustrate the gravity of public and patient interest in pharmacogenomic testing. In this case, the breast-feeding mother had taken codeine regularly for about 14 days when her 13-day-old infant died from toxic levels of morphine. Unknown to her and the prescriber, both the mother and infant were ultra-rapid CYP2D6 metabolizers, resulting in a more rapid and extensive conversion of codeine to morphine.
A logical strategy for preventing similar deaths would be routine CYP2D6 genotyping when prescribing codeine to breast-feeding mothers. However, after several investigations examined the metabolic and excretion pathways of codeine in their entirety, the FDA did not recommend routine CYP2D6 testing when prescribing codeine to breastfeeding mothers because several other factors, including rare genetic variations of other enzymes, proved necessary for reaching the opioid toxicity leading to the infant’s death.62
PHARMACOGENOMICS OF PSYCHOTROPIC DRUGS
Pharmacogenomic testing has clinical utility for some psychotropic drugs.
HLA-B and carbamazepine
Considered a standard of care, HLA-B genotyping is appropriate before prescribing carbamazepine (Tegretol, Equetro) to patients in populations in which HLAB*1502 is likely to be present, such as Asians. Carriers of HLAB* 1502 are at higher risk of life-threatening skin reactions such as Stevens-Johnson syndrome.11
Several other pharmacogenomic applications for psychotropic medications have been suggested, but routine testing has not been recommended by the FDA or endorsed by any expert panel because sufficient clinical utility and cost-effectiveness have not been demonstrated. A brief summary of study findings and a few practical suggestions follow.
Polymorphisms in metabolizing enzymes have been investigated in patients receiving psychotropic drugs.
CYP2D6 and antidepressants
Many antidepressants show significant differences in plasma drug levels with CYP2D6 polymorphisms (in descending order of influence)55:
- Imipramine (Tofranil)
- Doxepin (Adapin, Silenor, Sinequan)
- Maprotiline (Deprilept, Ludiomil, Psymion)
- Trimipramine (Surmontil)
- Desipramine (Noraprim)
- Nortriptyline (Aventyl, Pamelor)
- Clomipramine (Anafranil)
- Paroxetine (Paxil)
- Venlafaxine (Effexor)
- Amitriptyline (Elavil)
- Mianserin
- Trazadone (Desyrel)
- Bupropion (Wellbutrin)
- Nefazodone (Serzone)
- Citalopram (Celexa)
- Sertraline (Zoloft).
CYP2D6 and antipsychotics
Several antipsychotics are also influenced by CYP2D6 polymorphisms (also in descending order of influence)55:
- Perphenazine (Trilafon)
- Thioridazine (Mellaril)
- Olanzapine (Zyprexa)
- Zuclopenthixol (Cisordinol, Clopixol, Acuphase)
- Aripiprazole (Abilify)
- Flupentixol (Depixol, Fluanxol)
- Haloperidol (Haldol)
- Perazine (Taxilan)
- Risperidone (Risperdal)
- Pimozide (Orap).
CYP2C19 and antidepressants
CYP2C19 polymorphisms are likewise associated with differences in drug metabolism for many antidepressants, such as (in descending order of CYP2C19-mediated influence)55:
- Trimipramine
- Doxepin
- Amitriptyline
- Imipramine
- Citalopram (Celexa)
- Clomipramine
- Moclobemide (Aurorix, Manerix)
- Sertraline
- Fluvoxamine (Luvox).
Clinical relevance of CYP2D6 and CYP2C19
Several studies have demonstrated that poor and intermediate CYP2D6 metabolizers have a higher incidence of adverse effects when taking CYP2D6-dependent antidepressants63–68; however, an almost equal number of studies did not find statistically significant associations.69–72 Likewise, several studies have found an association between ultra-rapid CYP2D6 metabolizer status and diminished response to antidepressants,65,73,74 but no association was found in a larger retrospective study.75
Routine CYP2D6 and CYP2C19 screening is not recommended when prescribing psychotropic drugs. However, reviews of the pharmacokinetic data have suggested a few practical applications when genetic status is already known. In general, clinicians can consider reducing the dose of tricyclic antidepressants by about 50% when prescribing to CYP2D6-poor-metabolizers.55,76–78
Genes that affect serotonin metabolism
Several genes in the serotonin pathway have been investigated to determine whether they influence patients’ susceptibility to depression and adverse effects and response to psychotropic medications.
SLC6A4. Polymorphisms in the promoter region of the serotonin transporter gene SLC6A4 appear to influence the treatment response and side-effect profiles of selective serotonin reuptake inhibitors (SSRIs). Carriers of the SLC6A4 5-HTTLPR L alleles have fewer side effects79 and better response to SSRI treatment, and carriers of the S allele have a higher incidence of antidepressant-induced mania80 and poorer response to SSRI treatment.81
5-HT. Polymorphisms in serotonin receptors (2A and 2C subtypes) appear to influence SSRI response and side effects. Carriers of 5-HT 2A C alleles had more severe adverse effects from paroxetine,71 but another 5-HT 2A polymorphism common to Asians is associated with better response to antidepressant therapy.82 A 5-HT 2C polymorphism was associated with a lower incidence of antipsychotic-induced weight gain.83
Although the understanding of these relationships is incomplete and routine pharmacogenomic testing is not currently recommended, reviews of the pharmacodynamic data have suggested a few practical applications when a patient’s genetic status is already known. One should consider:
- Selecting treatments other than SSRIs for depressed patients known to possess the SLC6A4 variant
- Selecting citalopram for depressed patients known to carry the 5-HT 2A polymorphism
- Avoiding treatment with antipsychotic drugs for patients known to possess the 5-HT 2C polymorphism.
THE FUTURE OF PHARMACOGENOMIC TESTING
The examples discussed in this article provide some insight about how pharmacogenomic testing is maturing and slowly being integrated into the practice of medicine. They also illustrate the complexity of the multiple stages of research that pharmacogenomic applications must go through in order to be adopted as standard practice.
In the future, pharmacogenomic data will continue to accumulate, and the clinical utility of many other pharmacogenomic tests may be uncovered. The FDA provides information on emerging pharmacogenomic tests at its Web site, www.fda.gov.11 Its up-to-date “Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels” includes boxed warnings, recommendations, research outcomes, and relevant population genetics.
If the FDA continues its current policy, prospective randomized trials that show improvement in patient outcomes will remain the gold standard for determining the clinical significance of a pharmacogenomic test. Furthermore, cost-benefit analyses are likely to continue dictating policy regarding pharmacogenomic testing, and cost-benefit profiles should improve as technology advances and as information gathered from a single test becomes applicable to multiple medications and clinical scenarios.
In the meantime, physicians should become familiar with the terms used in medical genetics and pharmacogenomics and begin to understand genetic contributions to the outcomes of drug therapy. For example, understanding the consequences of metabolizer status and the frequency of variants in a given population can be tremendously helpful when advising our patients about anticipating potential problems when taking specific medications and about making informed decisions about pharmacogenomic testing.
This exchange of information alone may go a long way in improving therapy outcomes even when prospective pharmacogenomic testing is not routinely performed. Furthermore, an increasing number of patients will already have genotyping information available when they come to us, and clinicians need to be aware of the many pharmacogenomic applications recommended by the FDA when genetic status is known.10
In many patients, certain drugs do not work as well as expected, whereas in other patients they cause toxic effects, even at lower doses. For some patients, the reason may be genetic.
Sizeable minorities of the population carry genetic variants—polymorphisms— that affect their response to various drugs. Thanks to genetic research, our understanding of the variability of drug response has advanced markedly in the last decade. Many relevant polymorphisms have been identified, and tests for some of them are available.
Armed with the knowledge of their patients’ genetic status, physicians could predict their response to certain drugs, leading to better efficacy, fewer adverse drug reactions, and a better cost-benefit ratio.
The possible impact is substantial, since more than half of the drugs most commonly involved in adverse drug reactions are metabolized by polymorphic enzymes.1 Adverse drug reactions remain a significant detriment to public health, having a substantial impact on rates of morbidity and death and on healthcare costs.2–8 In the United States, adverse drug reactions are a leading cause of death in hospitalized patients4 and are annually responsible for hundreds of thousands of deaths and hundreds of billions of dollars in added costs.2,4,6–8
In the meantime, physicians can educate their patients and promote efforts to incorporate genomic information into standard clinical decision-making.
This article offers an overview of pharmacogenomic testing, discussing implications and limitations of a few validated tests. Specifically, we will discuss testing that is relevant when using warfarin (Coumadin), clopidogrel (Plavix), statins, tamoxifen (Nolvadex), codeine, and psychotropic medications, as well as the future role of pharmacogenomic testing in medicine.
WHAT IS PHARMACOGENOMICS?
Pharmacogenomics is the study of how genetic factors relate to interindividual variability of drug response.
Many clinicians may not be familiar with the background and terminology used in the pharmacogenomic literature. Below, a brief review of the terminology is followed by a schematic describing the various stages of research involved in pharmacogenomics and the advancement of a test into standard practice.
The review and schematic may be helpful for evaluating the clinical significance of pharmacogenomics-related articles.
From genotype to phenotype
Genotype refers to the coding sequence of DNA base pairs for a particular gene, and phenotype (eg, disease or drug response) refers to a trait resulting from the protein product encoded by the gene. The name of a gene often refers to its protein product and is italicized (eg, the CYP3A4 gene encodes for the CYP3A4 enzyme).
Two alleles per autosomal gene (one paternal and one maternal) form the genotype. Heterozygotes possess two different alleles, and homozygotes possess two of the same alleles. The most common allele in a population is referred to as the wild type, and allele frequencies can vary greatly in different populations.9
Most sequence variations are single nucleotide polymorphisms (SNPs, pronounced “snips”), a single DNA base pair substitution that may result in a different gene product. SNPs can be classified as structural RNA polymorphisms (srSNPs), regulatory polymorphisms (rSNPs), or polymorphisms in coding regions (cSNPs)10: srSNPs alter mRNA processing and translation, rSNPs alter transcription, and cSNPs alter protein sequence and function.
Recently, genetic associations with a phenotype have been done on a large scale, with millions of SNPs measured in each of many subjects. This approach, called a genomewide association study or GWAS, has revealed countless candidate genes for clinical traits, but only a few have resulted in a practical clinical application. SNPs may by themselves exert a pharmacokinetic effect (ie, how the body processes the drug), a pharmacodynamic effect (ie, how the drug affects the body), or both, or they may act in concert with other genetic factors. Pharmacodynamic effects can result from a pharmacokinetic effect or can result from variations in a pharmacologic target.
Establishing a genotype-phenotype association can involve clinical studies, animal transgenic studies, or molecular and cellular functional assays.
Clinical applications are emerging
WARFARIN: IMPORTANCE OF CYP2C9, VKORC1
Warfarin is used for the long-term treatment and prevention of thromboembolic events.
This drug has a narrow therapeutic window and shows substantial interpatient dose variability. The start of warfarin therapy is associated with one of the highest rates of adverse events and emergency room visits of any single drug.12 More than 2 million patients start warfarin each year in the United States alone,13 and about 20% of them are hospitalized within the first 6 months because of bleeding due to overanticoagulation.14
The findings from a recent study suggest that pharmacogenomic testing may eventually allow more patients to safely benefit from warfarin therapy. In this large, nationwide, prospective study, hospitalization rates were 30% lower when pharmacogenomic testing was used.14 However, no reduction was seen in the time needed to reach the target international normalized ratio (INR) or in the need for INR checks at 6 months. Furthermore, this study used historical control data, and some or all of the reduction in hospitalization rates may be attributed to more frequent INR checks in the patients who underwent testing than in the historical control group.
A relationship between warfarin dose requirements and the genetic status of CYP2C9, which encodes a major drug-metabolizing enzyme, has been demonstrated in retrospective and prospective studies.15–17
S-warfarin is metabolized by CYP2C9, which is polymorphic
Warfarin contains equal amounts of two isomers, designated S and R. S-warfarin, which is more potent, is metabolized principally by CYP2C9, while R-warfarin is metabolized by CYP1A2, CYP2C19, and CYP3A4.
People who possess two copies of the wild type CYP2C9 gene CYP2C9*1 metabolize warfarin very well and so are called “extensive warfarin metabolizers.” Carriers of the allelic variants CYP2C9*2 and CYP2C9*3 (which have point mutations in exons 3 and 7 of CYP2C9, respectively), have less capacity. Compared with those who are homozygous for the wild-type gene, homozygous carriers of CYP2C9* 3 clear S-warfarin at a rate that is 90% lower, and those with the CYP2C9*1/*3, CYP2C9* 1/*2, CYP2C9*2/*2, or CYP2C9*2/*3 genotypes clear it at a rate 50% to 75% lower. A meta-analysis of 12 studies found that the CYP2C9 genotype accounted for 12% of the interindividual variability of warfarin dose requirements.18
About 8% of whites carry at least one copy of CYP2C9*2, as do 1% of African Americans; the allele is rare in Asian populations. The frequency of CYP2C9*3 is 6% in whites, 1% in African Americans, and 3% in Asians.19,20 People with CYP2C9*4 or CYP2C9*5 have a diminished capacity to clear warfarin; however, these variants occur so infrequently that their clinical relevance may be minimal.
Warfarin’s target, VKOR, is also polymorphic
Genetic variation in warfarin’s pharmacologic target, vitamin K 2,3-epoxide reductase (VKOR), also influences dose requirements. Warfarin decreases the synthesis of vitamin-K-dependent clotting factors by inhibiting VKOR. This inhibition depends on the patient’s C1 subunit gene, VKORC1. Patients with a guanine-to-adenine SNP 1,639 bases upstream of VKORC1 (−1639G>A) need lower warfarin doses—an average of 25% lower in those with the GA genotype (ie, one allele has guanine in the −1639 position and the other allele has adenine in that position) and 50% lower in those with the AA genotype compared with the wild-type genotype GG.21 This promoter SNP, positioned upstream (ie, before the gene-coding region), greatly influences VKORC1 expression.
A meta-analysis of 10 studies found that the VKORC1 polymorphism accounts for 25% of the interindividual variation in warfarin dose.18 In one study, the frequency of the AA genotype in a white population was 14%, AG 47%, and GG 39%; in a Chinese population the frequency of AA was 82%, AG 18%, and GG 0.35%.22
CYP4F2 and GGCX also affect warfarin’s dose requirements
Genetic variations in the enzymes CYP4F2 and gamma-glutamyl carboxylase (GGCX) also influence warfarin dose requirements. Although the data are limited and the effects are smaller than those of CYP2C9 and VKORC1, people with a SNP in CYP4F2 need 8% higher doses of warfarin, while those with a SNP in GGCX need 6% lower doses.23
CYP2C9 and VKORC1 testing is available
Currently, the clinical pharmacogenetic tests relevant for warfarin use are for CYP2C9 and VKORC1.10
The FDA has approved four warfarin pharmacogenetic test kits, but most third-party payers are reluctant to reimburse for testing because it is not currently considered a standard of care. Testing typically costs a few hundred dollars, but it should become less expensive as it becomes more commonplace. The current FDA-approved product label for warfarin does not recommend routine pharmacogenomic testing for determining initial or maintenance doses, but it does acknowledge that dose requirements are influenced by CYP2C9 and VKORC1 and states that genotype information, when available, can assist in selecting the starting dose.24
Clinical trials of warfarin pharmacogenomic testing are under way
Although genetic status can greatly influence an individual patient’s warfarin dosing requirement, routine prospective pharmacogenomic testing is not endorsed by the FDA or by other expert panels26 because there is currently insufficient evidence to recommend for or against it.
Several large prospective trials are under way. For example, the National Heart, Lung, and Blood Institute began a prospective trial in about 1,200 patients to evaluate the use of clinical plus genetic information to guide the initiation of warfarin therapy and to improve anticoagulation control for patients.27 The results, expected in September 2011, and those of other large prospective trials should provide adequate evidence for making recommendations about the clinical utility of routine pharmacogenetic testing for guiding warfarin therapy.
Several recent cost-utility and cost-effectiveness studies have attempted to quantify the value of pharmacogenomic testing for warfarin therapy,28–30 but their analyses are largely limited because the benefit (clinical utility) is yet to be sufficiently characterized.
The relevance of such analyses may soon be drastically diminished, as several non-vitamin-K-dependent blood thinners such as rivaroxaban (Xarelto), dabigatran (Pradaxa), and apixaban are poised to enter clinical practice.31
CLOPIDOGREL IS ACTIVATED BY CYP2C19
Clopidogrel, taken by about 40 million patients worldwide, is used to prevent atherothrombotic events and cardiac stent thrombosis when given along with aspirin.
Studies of clopidogrel pharmacogenomics
A recent genome-wide association study conducted in a cohort of 429 healthy Amish persons revealed a SNP in CYP2C19 to be associated with a diminished response to clopidogrel and to account for 12% of the variation in drug response.33 Traditional factors (the patient’s age, body-mass index, and cholesterol level) combined accounted for less than 10% of the variation.
Findings were similar in a subsequent investigation in 227 cardiac patients receiving clopidogrel: 21% of those with the variant had a cardiovascular ischemic event or died during a 1-year follow-up period compared with 10% of those without the variant (hazard ratio 2.42, P = .02).33
A 12-year prospective study investigating clopidogrel efficacy in 300 cardiac patients under the age of 45 used cardiovascular death, nonfatal myocardial infarction, and urgent coronary revascularization as end points. It concluded that the only independent predictor of these events was the patient’s CYP2C19 status.34
A study in 2,200 patients with recent myocardial infarction examined whether any of the known allelic variations that modulate clopidogrel’s absorption (ABCB1), metabolic activation (CYP3A4/5 and CYP2C19), or biologic activity (P2RY12 and ITGB3) was associated with a higher rate of the combined end point of all-cause mortality, nonfatal myocardial infarction, or stroke. None of the SNPs in CYP3A4/5, P2RY12, or ITGB3 that were evaluated was associated with a higher risk at 1 year. However, the allelic variations modulating clopidogrel’s absorption (ABCB1) and metabolism (CYP2C19) were associated with higher event rates. Patients with two variant ABCB1 alleles had a higher adjusted hazard ratio (95% confidence interval [CI] 1.2–2.47) than those with the wild-type allele. Patients who had one or two CYP2C19 loss-of-function alleles had a higher event rate than those with two wild-type alleles (95% CI 1.10–3.58 and 1.71–7.51, respectively).35
Conversely, researchers from the Population Health Research Institute found no association between poor-metabolizer status and treatment outcomes when CYP2C19 analysis was retrospectively added to the findings of two large clinical trials (combined N > 5,000). However, patients with acute coronary syndrome benefited more from clopidogrel treatment if they were ultra-rapid metabolizers (possessing the gain-of-function allele CYP2C19*17).36
Current status of clopidogrel testing: Uncertain
A current FDA boxed warning states that poor CYP2C19 metabolizers may not benefit from clopidogrel and recommends that prescribers consider alternative treatment for patients in this category.37 However, routine CYP2C19 testing is not recommended, and no firm recommendations have been established regarding dose adjustments for CYP2C19 status.
In 2010, the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association collectively pronounced the current evidence base insufficient for recommending routine pharmacogenomic testing.39
Needed are large-scale studies examining the cost-effectiveness and clinical utility of genotype-guided clopidogrel therapy compared with other therapy options such as prasugrel (Effient), an analogue not metabolized by CYP2C19. One such study, sponsored by Medco Health Solutions, plans to enroll 14,600 cardiac patients and has an estimated completion date in June 2011.40 The expectation that clopidogrel will be available in generic form in 2012 adds to the uncertainty regarding the cost-effectiveness of CYP2C19 testing for clopidogrel therapy.
STATINS: SLC01B1*5 INCREASES MYOPATHY RISK
Statins lower the concentration of low-density lipoprotein cholesterol (LDL-C), resulting in a relative-risk reduction of about 20% for each 1 mmol/L (39 mg/dL) decrement in LDL-C.41 They are one of the most commonly prescribed classes of drugs, but their side effects can limit their appeal: statin-induced myopathy occurs in about 1:1,000 to 1:10,000 patients and is difficult to predict.
SLC01B1. The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH), a genome-wide association study, recently found a SNP (SLCO1B1* 5) in the SLC01B1 gene to be associated with a higher risk of statin-induced myopathy in cardiac patients receiving simvastatin (Zocor) 40 or 80 mg daily.42 The SLC01B1 gene, located on chromosome 12, influences the extent of the drug’s hepatic uptake and its serum concentration. Only the SLC01B1*5 SNP emerged as a predictor of statin-induced myopathy across the entire genome.42
The authors believe the findings are likely to apply to other statins. The mechanisms leading to statin-induced myopathy and the impact of statin pharmacogenomics are still unclear.43
CYP3A4. Other genetic variants may play a vital role in determining response to statin therapy. Carriers of a newly identified CYP3A4 polymorphism (intron 6 SNP, rs35599367, C>T) required significantly lower statin doses (0.2–0.6 times less) for optimal lipid control. The analyses included atorvastatin (Lipitor), simvastatin, and lovastatin (Mevacor), and the association was robust (P = .019).44
Statin pharmacogenomic testing is not routinely recommended
Routine pharmacogenomic testing for statin therapy is not recommended. Additional studies are needed to determine the clinical utility and cost-effectiveness of pharmacogenomic testing (involving a combination of several polymorphisms) in various patient populations delineated by type of statin, dose, and concomitant use of other drugs.
TAMOXIFEN IS ACTIVATED BY CYP2D6
Tamoxifen is prescribed to prevent the recurrence of estrogen-receptor-positive breast cancer, to treat metastatic breast cancer, to prevent cancer in high-risk populations, and to treat ductal carcinoma in situ.
Tamoxifen is metabolized to form endoxifen, which has much higher potency and higher systemic levels than tamoxifen.45 Both CYP2D6 and CYP3A4/5 are required to produce endoxifen via two intermediates, but CYP2D6 catalyzes the critical step leading to metabolic activation.
The CYP2D6 gene is highly polymorphic, with more than 75 allelic variants identified. Extensive literature is available describing the influence of CYP2D6 polymorphisms on tamoxifen metabolism and therapy outcomes.46–52 Several CYP2D6 variants result in reduced or no enzyme activity, and people who have more than two normally functioning alleles have exaggerated enzyme activity (gene amplification).
Classification of CYP2D6 status
Several systems have been developed to categorize the phenotypic activity of CYP2D6 based on genotype.
A genetic basis for the observed diversity in the metabolism of cytochrome P450 substrates was recognized more than 30 years ago. People were categorized as either extensive or poor metabolizers, reflecting normal vs impaired ability to metabolize the CYP2D6 substrates sparteine and debrisoquine. Later work expanded this system to include categories for intermediate (between poor and extensive) and ultra-rapid (better than extensive) metabolizers.
The genetic basis for these categories includes homozygosity for dysfunctional variants (the poor-metabolizer group) or extra copies of normal functioning variants (the ultra-rapid-metabolizer group).
Newer systems have been described for characterizing the CYP2D6 activity phenotype whereby CYP2D6 variants are assigned activity scores.53–56 The various scoring systems have been reviewed by Kirchheiner.57
A recent version of the activity scoring system also takes into consideration the many drugs that inhibit CYP2D6, such as amiodarone (Cordarone) and fluoxetine (Prozac) that can reduce the action of tamoxifen if given with it (Table 4).58 For example, the tamoxifen exposure (as predicted by the CYP2D6-activity score) experienced by a CYP2D6 extensive metabolizer taking a CYP2D6-inhibiting drug may be similar to the exposure experienced by a CYP2D6 poor metabolizer receiving the same tamoxifen dose but not taking a CYP2D6-inhibiting drug.
Likewise, the activity score of a CYP2D6 intermediate metabolizer taking a CYP2D6-inducing drug may be similar to that of a CYP2D6 ultra-rapid metabolizer not taking a CYP2D6-inducing drug. Examples of CYP2D6 inducers are dexamethasone, rifampin, and hyperforin (St. John’s wort).
While the newer systems are reported to provide better correlations between genotype and phenotype scores, the older scoring systems and the categorical names are still widely used (eg, in the FDA-approved AmpliChip CYP450 test from Roche,59 which includes genotype data for CYP2D6 and CYP2C19).
No firm recommendations for CYP2D6 testing in tamoxifen users
The different genotypes and phenotypes vary in prevalence in different ethnic groups, and significantly different activity levels for endoxifen formation are observed. Tamoxifen lacks efficacy in those who are poor CYP2D6 metabolizers—ie, about 7% of the white population.
However, the FDA has not made firm recommendations about CYP2D6 testing for prescribing tamoxifen because the evidence of benefit, although suggestive, has been considered insufficient.
Clinicians should be aware that tamoxifen’s efficacy is greatly reduced by concomitant therapy with CYP2D6-inhibiting drugs (Table 4).
Other genes affecting tamoxifen: CYP3A4/5, SULT1A1, and UGT2B15
Some investigators propose that polymorphisms in additional genes encoding enzymes in the tamoxifen metabolic and elimination pathways (eg, CYP3A4/5, SULT1A1, and UGT2B15) also need to be considered to account adequately for interindividual variation in drug response.
For example, CYP3A4 and CYP3A5 are also polymorphic, and large interindividual variation exists in their enzyme activities. These enzymes have overlapping substrate specificities, represent the most abundant drug-metabolizing enzymes in the human liver, and are involved in the biotransformation of a broad range of endogenous substrates and most drugs.60
Clinical studies evaluating the impact of CYP3A4/5 polymorphisms have been inconsistent in their conclusions, which is generally attributed to the relatively low functional impact or the low prevalence of the SNPs evaluated. Many of the nearly 100 CYP3A4/5 polymorphisms identified have not yet been characterized regarding their functional impact on enzyme expression or activity. CYP-3A4/5 enzyme activity is highly variable between individuals and warrants further study of its role in outcomes of tamoxifen therapy. Ongoing and future prospective clinical trials evaluating CYP2D6, CYP3A4/5, and other relevant polymorphisms are necessary to define their clinical relevance before routine genetic testing for tamoxifen can be justified.
CODEINE IS ALSO ACTIVATED BY CYP2D6
Codeine also depends on the CYP2D6 gene, as it must be activated to its more potent opioid metabolites, including morphine. Poor CYP2D6 metabolizers do not benefit from codeine therapy.
The pharmacogenomics of codeine has become a hot topic, especially regarding breast-feeding mothers. The debate was ignited with the publication in 2006 of a case report of an infant’s death, apparently the result of metabolic polymorphisms.61 The evolution of this debate and the outcome of the case may be noteworthy to clinicians, as they illustrate the gravity of public and patient interest in pharmacogenomic testing. In this case, the breast-feeding mother had taken codeine regularly for about 14 days when her 13-day-old infant died from toxic levels of morphine. Unknown to her and the prescriber, both the mother and infant were ultra-rapid CYP2D6 metabolizers, resulting in a more rapid and extensive conversion of codeine to morphine.
A logical strategy for preventing similar deaths would be routine CYP2D6 genotyping when prescribing codeine to breast-feeding mothers. However, after several investigations examined the metabolic and excretion pathways of codeine in their entirety, the FDA did not recommend routine CYP2D6 testing when prescribing codeine to breastfeeding mothers because several other factors, including rare genetic variations of other enzymes, proved necessary for reaching the opioid toxicity leading to the infant’s death.62
PHARMACOGENOMICS OF PSYCHOTROPIC DRUGS
Pharmacogenomic testing has clinical utility for some psychotropic drugs.
HLA-B and carbamazepine
Considered a standard of care, HLA-B genotyping is appropriate before prescribing carbamazepine (Tegretol, Equetro) to patients in populations in which HLAB*1502 is likely to be present, such as Asians. Carriers of HLAB* 1502 are at higher risk of life-threatening skin reactions such as Stevens-Johnson syndrome.11
Several other pharmacogenomic applications for psychotropic medications have been suggested, but routine testing has not been recommended by the FDA or endorsed by any expert panel because sufficient clinical utility and cost-effectiveness have not been demonstrated. A brief summary of study findings and a few practical suggestions follow.
Polymorphisms in metabolizing enzymes have been investigated in patients receiving psychotropic drugs.
CYP2D6 and antidepressants
Many antidepressants show significant differences in plasma drug levels with CYP2D6 polymorphisms (in descending order of influence)55:
- Imipramine (Tofranil)
- Doxepin (Adapin, Silenor, Sinequan)
- Maprotiline (Deprilept, Ludiomil, Psymion)
- Trimipramine (Surmontil)
- Desipramine (Noraprim)
- Nortriptyline (Aventyl, Pamelor)
- Clomipramine (Anafranil)
- Paroxetine (Paxil)
- Venlafaxine (Effexor)
- Amitriptyline (Elavil)
- Mianserin
- Trazadone (Desyrel)
- Bupropion (Wellbutrin)
- Nefazodone (Serzone)
- Citalopram (Celexa)
- Sertraline (Zoloft).
CYP2D6 and antipsychotics
Several antipsychotics are also influenced by CYP2D6 polymorphisms (also in descending order of influence)55:
- Perphenazine (Trilafon)
- Thioridazine (Mellaril)
- Olanzapine (Zyprexa)
- Zuclopenthixol (Cisordinol, Clopixol, Acuphase)
- Aripiprazole (Abilify)
- Flupentixol (Depixol, Fluanxol)
- Haloperidol (Haldol)
- Perazine (Taxilan)
- Risperidone (Risperdal)
- Pimozide (Orap).
CYP2C19 and antidepressants
CYP2C19 polymorphisms are likewise associated with differences in drug metabolism for many antidepressants, such as (in descending order of CYP2C19-mediated influence)55:
- Trimipramine
- Doxepin
- Amitriptyline
- Imipramine
- Citalopram (Celexa)
- Clomipramine
- Moclobemide (Aurorix, Manerix)
- Sertraline
- Fluvoxamine (Luvox).
Clinical relevance of CYP2D6 and CYP2C19
Several studies have demonstrated that poor and intermediate CYP2D6 metabolizers have a higher incidence of adverse effects when taking CYP2D6-dependent antidepressants63–68; however, an almost equal number of studies did not find statistically significant associations.69–72 Likewise, several studies have found an association between ultra-rapid CYP2D6 metabolizer status and diminished response to antidepressants,65,73,74 but no association was found in a larger retrospective study.75
Routine CYP2D6 and CYP2C19 screening is not recommended when prescribing psychotropic drugs. However, reviews of the pharmacokinetic data have suggested a few practical applications when genetic status is already known. In general, clinicians can consider reducing the dose of tricyclic antidepressants by about 50% when prescribing to CYP2D6-poor-metabolizers.55,76–78
Genes that affect serotonin metabolism
Several genes in the serotonin pathway have been investigated to determine whether they influence patients’ susceptibility to depression and adverse effects and response to psychotropic medications.
SLC6A4. Polymorphisms in the promoter region of the serotonin transporter gene SLC6A4 appear to influence the treatment response and side-effect profiles of selective serotonin reuptake inhibitors (SSRIs). Carriers of the SLC6A4 5-HTTLPR L alleles have fewer side effects79 and better response to SSRI treatment, and carriers of the S allele have a higher incidence of antidepressant-induced mania80 and poorer response to SSRI treatment.81
5-HT. Polymorphisms in serotonin receptors (2A and 2C subtypes) appear to influence SSRI response and side effects. Carriers of 5-HT 2A C alleles had more severe adverse effects from paroxetine,71 but another 5-HT 2A polymorphism common to Asians is associated with better response to antidepressant therapy.82 A 5-HT 2C polymorphism was associated with a lower incidence of antipsychotic-induced weight gain.83
Although the understanding of these relationships is incomplete and routine pharmacogenomic testing is not currently recommended, reviews of the pharmacodynamic data have suggested a few practical applications when a patient’s genetic status is already known. One should consider:
- Selecting treatments other than SSRIs for depressed patients known to possess the SLC6A4 variant
- Selecting citalopram for depressed patients known to carry the 5-HT 2A polymorphism
- Avoiding treatment with antipsychotic drugs for patients known to possess the 5-HT 2C polymorphism.
THE FUTURE OF PHARMACOGENOMIC TESTING
The examples discussed in this article provide some insight about how pharmacogenomic testing is maturing and slowly being integrated into the practice of medicine. They also illustrate the complexity of the multiple stages of research that pharmacogenomic applications must go through in order to be adopted as standard practice.
In the future, pharmacogenomic data will continue to accumulate, and the clinical utility of many other pharmacogenomic tests may be uncovered. The FDA provides information on emerging pharmacogenomic tests at its Web site, www.fda.gov.11 Its up-to-date “Table of Valid Genomic Biomarkers in the Context of Approved Drug Labels” includes boxed warnings, recommendations, research outcomes, and relevant population genetics.
If the FDA continues its current policy, prospective randomized trials that show improvement in patient outcomes will remain the gold standard for determining the clinical significance of a pharmacogenomic test. Furthermore, cost-benefit analyses are likely to continue dictating policy regarding pharmacogenomic testing, and cost-benefit profiles should improve as technology advances and as information gathered from a single test becomes applicable to multiple medications and clinical scenarios.
In the meantime, physicians should become familiar with the terms used in medical genetics and pharmacogenomics and begin to understand genetic contributions to the outcomes of drug therapy. For example, understanding the consequences of metabolizer status and the frequency of variants in a given population can be tremendously helpful when advising our patients about anticipating potential problems when taking specific medications and about making informed decisions about pharmacogenomic testing.
This exchange of information alone may go a long way in improving therapy outcomes even when prospective pharmacogenomic testing is not routinely performed. Furthermore, an increasing number of patients will already have genotyping information available when they come to us, and clinicians need to be aware of the many pharmacogenomic applications recommended by the FDA when genetic status is known.10
- King HC, Sinha AA. Gene expression profile analysis by DNA micro-arrays: promise and pitfalls. JAMA 2001; 286:2280–2288.
- Nuckols TK, Paddock SM, Bower AG, et al. Costs of intravenous adverse drug events in academic and nonacademic intensive care units. Med Care 2008; 46:17–24.
- Gurwitz JH, Field TS, Judge J, et al. The incidence of adverse events in two long-term care facilities. Am J Med 2005; 118:251–258.
- Vargas E, Terleira A, Hernando F, et al. Effects of adverse drug reactions on length of stay in surgical intensive care units. Crit Care Med 2003; 31:694–698.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998; 279:1200–1205.
- Field TS, Gilman BH, Subramanian S, Fuller JC, Bates DW, Gurwitz JH. The costs associated with adverse drug events in older adults in the ambulatory setting. Med Care 2005; 43:1171–1176.
- Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA 1997; 277:301–306.
- Ernst FR, Grizzle AJ. Drug-related morbidity and mortality: updating the cost-of-illness model. J Am Pharm Assoc (Wash) 2001; 41:192–199.
- Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002; 3:229–243.
- Sadee W. Measuring cis-acting regulatory variants genome-wide: new insights into expression genetics and disease susceptibility. Genome Med 2009; 1:116.
- US Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labels. http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm. Accessed 1/18/2011.
- Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med 2007; 147:755–765.
- Elias DJ, Topol EJ. Warfarin pharmacogenomics: a big step forward for individualized medicine: enlightened dosing of warfarin. Eur J Hum Genet 2008; 16:532–534.
- Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study). J Am Coll Cardiol 2010; 55:2804–2812.
- Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 1994; 4:39–42.
- Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002; 287:1690–1698.
- Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med 2005; 7:97–104.
- Au N, Rettie AE. Pharmacogenomics of 4-hydroxycoumarin anticoagulants. Drug Metab Rev 2008; 40:355–375.
- García-Martín E, Martínez C, Ladero JM, Agúndez JA. Interethnic and intraethnic variability of CYP2C8 and CYP2C9 polymorphisms in healthy individuals. Mol Diagn Ther 2006; 10:29–40.
- Sullivan-Klose TH, Ghanayem BI, Bell DA, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics 1996; 6:341–349.
- Wen MS, Lee M, Chen JJ, et al. Prospective study of warfarin dosage requirements based on CYP2C9 and VKORC1 genotypes. Clin Pharmacol Ther 2008; 84:83–89.
- Larramendy-Gozalo C, Yang JQ, Verstuyft C, et al. Genetic polymorphism of vitamin K epoxide reductase (VKORC1) 1173C>T in a Chinese and a Caucasian population. Basic Clin Pharmacol Toxicol 2006; 98:611–613.
- Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood 2008; 111:4106–4112.
- Bristol-Myers Squibb. Coumadin (warfarin sodium) Prescribing Information. January 2010.
- Barnes-Jewish Hospital at Washington University Medical Center. Warfarin dosing. http://warfarindosing.orgAccessed 1/20/2011.
- Flockhart DA, O’Kane D, Williams MS, et al. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med 2008; 10:139–150.
- National Institutes of Health. ClinicalTrials.gov. http://clinicaltrials.gov. Accessed January 20, 2011.
- Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med 2009; 150:73–83.
- Meckley LM, Gudgeon JM, Anderson JL, Williams MS, Veenstra DL. A policy model to evaluate the benefits, risks, and costs of warfarin pharmacogenomic testing. Pharmacoeconomics 2010; 28:61–74.
- Patrick AR, Avron J, Choudhry NK. Cost-effectiveness of genotype-guided warfarin dosing for patients with atrial fibrillation. Circ Cardiovasc Qual Outcomes 2009; 2:429–436.
- Haas S. New oral Xa and IIa inhibitors: updates on clinical trial results. J Thromb Thrombolysis 2008; 25:52–60.
- Bhatt DL. Tailoring antiplatelet therapy based on pharmacogenomics: how well do the data fit? JAMA 2009; 302:896–897.
- Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009; 302:849–857.
- Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 2009; 373:309–317.
- Simon T, Verstuyft C, Mary-Krause M, et al; French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009; 360:363–375.
- Paré G, Mehta SR, Yusuf S, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med 2010; 363:1704–1714.
- Bristol-Myers Squibb/Sanofi Pharmaceutical Partnership. Plavix (clopidogrel bisulfate) prescribing information. August 2010.
- P450 Drug Interaction Table. Indiana University School of Medicine. http://medicine.iupui.edu/clinpharm/ddis/table.asp. Accessed 1/21/2011.
- Society for Cardiovascular Angiography and Interventions; Holmes DR, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation 2010; 122:537–557.
- National Institutes of Health. Genotype Guided Comparison of Clopidogrel and Prasugrel Outcomes Study. http://clinicaltrialsfeeds.org/clinical-trials/show/NCT00995514. Accessed 1/20/2011.
- Amarenco P, Labreuche J. Lipid management in the prevention of stroke: Review and updated meta-analysis of statins for stroke prevention. Lancet Neurol 2009; 8:453–463.
- SEARCH Collaborative Group, Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med 2008; 359:789–799.
- Romaine SP, Bailey KM, Hall AS, Balmforth AJ. The influence of SLC01B1 (OATP1B1) gene polymorphisms on response to statin therapy. Pharmacogenomics J 2010; 10:1–11.
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 2010; Apr 13 [Epub ahead of print].
- Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 2005; 23:9312–9318.
- Beverage JN, Sissung TM, Sion AM, Danesi R, Figg WD. CYP2D6 polymorphisms and the impact on tamoxifen therapy. J Pharm Sci 2007; 96:2224–2231.
- Flockhart D. CYP2D6 genotyping and the pharmacogenetics of tamoxifen. Clin Adv Hematol Oncol 2008; 6:493–494.
- Goetz MP, Kamal A, Ames MM. Tamoxifen pharmacogenomics: the role of CYP2D6 as a predictor of drug response. Clin Pharmacol Ther 2008; 83:160–166.
- Stearns V, Rae JM. Pharmacogenetics and breast cancer endocrine therapy: CYP2D6 as a predictive factor for tamoxifen metabolism and drug response? Expert Rev Mol Med 2008; 10:e34.
- Dezentjé VO, Guchelaar HJ, Nortier JW, van del Velde CJ, Gelderblom H. Clinical implications of CYP2D6 genotyping in tamoxifen treatment for breast cancer. Clin Cancer Res 2009; 15:15–21.
- Higgins MJ, Rae JM, Flockhart DA, Hayes DF, Stearns V. Pharmacogenetics of tamoxifen: who should undergo CYP2D6 genetic testing? J Natl Compr Canc Netw 2009; 7:203–213.
- Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer 2009; 9:576–586.
- Steimer W, Zöpf K, von Amelunxen S, et al. Allele-specific change of concentration and functional gene dose for the prediction of steady-state serum concentrations of amitriptyline and nortriptyline in CYP2C19 and CYP2D6 extensive and intermediate metabolizers. Clinical Cancer 2004; 50:1623–1633.
- Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharm Ther 2008; 83:234–242.
- Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry 2004; 9:442–473.
- Kirchheiner J, Schmidt H, Tzvetkov M, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J 2007; 7:257–265.
- Kirchheiner J. CYP2D6 phenotype prediction from genotype: which system is the best? Clin Pharmacol Ther 2008; 83:225–227.
- Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol 2010; 50:450–458.
- Hoffmann-La Roche Ltd. AmpliChip CYP450 Test. http://www.roche.com/assays/Pages/AmpliChipCYP450Test.aspx. Accessed 1/21/2011.
- Anzenbacher P, Anzenbacherová E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci 2001; 58:737–747.
- Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet 2006; 368:704.
- Willmann S, Edginton AN, Coboeken K, Ahr G, Lippert J. Risk to the breast-fed neonate from codeine treatment to the mother: a quantitative mechanistic modeling study. Clin Pharm Ther 2009; 86:634–643.
- Grzesiak M, Beszłej A, Lebioda A, Jonkisz A, Dobosz T, Kienja A. [Retrospective assessment of the antidepressants tolerance in the group of patients with diagnosis of depression and different CYP2D6 genotype.] [In Polish] Psychiatr Pol 2003; 37:433–444.
- Laika B, Leucht S, Heres S, Steimer W. Intermediate metabolizer: increased side effects in psychoactive drug therapy. The key to cost-effectiveness of pretreatment CYP2D6 screening? Pharmacogenomics J 2009; 9:395–403.
- Rau T, Wohlleben G, Wuttke H, et al. CYP2D6 genotype: Impact on adverse effects and nonresponse during treatment with antidepressants—a pilot study. Clin Pharm Ther 2004; 75:386–393.
- McAlpine DE, O’Kane DJ, Black JL, Mrazek DA. Cytochrome P450 2D6 genotype variation and venlafaxine dosage. Mayo Clin Proc 2007; 82:1065–1068.
- Chen S, Chou WH, Blouin RA, et al. The cytochrome P450 2D6 (CYP2D6) enzyme polymorphism: screening costs and influence on clinical outcomes in psychiatry. Clin Pharmacol Ther 1996; 60:522–534.
- Shams ME, Arneth B, Hiemke C, et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther 2006; 31:493–502.
- Whyte EM, Romkes M, Mulsant BH, et al. CYP2D6 genotype and venlafaxine-XR concentrations in depressed elderly. Int J Geriatr Psychiatry 2006; 21:542–549.
- Roberts RL, Mulder RT, Joyce PR, Luty SE, Kennedy MA. No evidence of increased adverse drug reactions in cytochrome P450 CYP2D6 poor metabolizers treated with fluoxetine or nortriptyline. Hum Psychopharmacol 2004; 19:17–23.
- Murphy GM, Kremer C, Rodrigues HE, Schatzberg AF. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry 2003; 160:1830–1835.
- Gillman PK. Re: no evidence of increased adverse drug reactions in cytochrome P450 CYP2D6 poor metabolizers treated with fluoxetine or nortriptyline. Hum Psychopharmacol 2005; 20:61–62.
- Gex-Fabry M, Eap CB, Oneda B, et al. CYP2D6 and ABCB1 genetic variability: influence on paroxetine plasma level and therapeutic response. Ther Drug Monit 2008; 30:474–482.
- Kawanishi C, Lundgren S, Agren H, Bertilsson L. Increased incidence of CYP2D6 gene duplication in patients with persistent mood disorders: ultrarapid metabolism of antidepressants as a cause of nonresponse. A pilot study. Eur J Clin Pharmacol 2004; 59:803–807.
- Serretti A, Calati R, Massat I, et al. Cytochrome P450 CYP1A2, CYP2C9, CYP2C19 and CYP2D6 genes are not associated with response and remission in a sample of depressive patients. Int Clin Psychopharmacol 2009; 24:250–256.
- de Leon J, Armstrong SC, Cozza KL. Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics 2006; 47:75–85.
- de Leon J, Susce MT, Johnson M, et al. DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr 2009; 14:19–34.
- Thuerauf N, Lunkenheimer J. The impact of the CYP2D6-polymorphism on dose recommendations for current antidepressants. Eur Arch Psychiatry Clin Neurosci 2006; 256:287–293.
- Horstmann S, Binder EB. Pharmacogenomics of antidepressant drugs. Pharmacol Ther 2009; 124:57–73.
- Ferreira Ade A, Neves FS, da Rocha FF, et al. The role of 5-HTTLPR polymorphism in antidepressant-associated mania in bipolar disorder. J Affect Disord 2009; 112:267–272.
- Serretti A, Kato M, De Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry 2007; 12:247–257.
- Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry 2010; 15:473–500.
- Reynolds GP, Zhang Z, Zhang X. Polymorphism of the promoter region of the serotonin 5-HT(2C) receptor gene and clozapine-induced weight gain. Am J Psychiatry 2003; 160:677–679.
- King HC, Sinha AA. Gene expression profile analysis by DNA micro-arrays: promise and pitfalls. JAMA 2001; 286:2280–2288.
- Nuckols TK, Paddock SM, Bower AG, et al. Costs of intravenous adverse drug events in academic and nonacademic intensive care units. Med Care 2008; 46:17–24.
- Gurwitz JH, Field TS, Judge J, et al. The incidence of adverse events in two long-term care facilities. Am J Med 2005; 118:251–258.
- Vargas E, Terleira A, Hernando F, et al. Effects of adverse drug reactions on length of stay in surgical intensive care units. Crit Care Med 2003; 31:694–698.
- Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998; 279:1200–1205.
- Field TS, Gilman BH, Subramanian S, Fuller JC, Bates DW, Gurwitz JH. The costs associated with adverse drug events in older adults in the ambulatory setting. Med Care 2005; 43:1171–1176.
- Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA 1997; 277:301–306.
- Ernst FR, Grizzle AJ. Drug-related morbidity and mortality: updating the cost-of-illness model. J Am Pharm Assoc (Wash) 2001; 41:192–199.
- Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002; 3:229–243.
- Sadee W. Measuring cis-acting regulatory variants genome-wide: new insights into expression genetics and disease susceptibility. Genome Med 2009; 1:116.
- US Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labels. http://www.fda.gov/Drugs/ScienceResearch/ResearchAreas/Pharmacogenetics/ucm083378.htm. Accessed 1/18/2011.
- Budnitz DS, Shehab N, Kegler SR, Richards CL. Medication use leading to emergency department visits for adverse drug events in older adults. Ann Intern Med 2007; 147:755–765.
- Elias DJ, Topol EJ. Warfarin pharmacogenomics: a big step forward for individualized medicine: enlightened dosing of warfarin. Eur J Hum Genet 2008; 16:532–534.
- Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study). J Am Coll Cardiol 2010; 55:2804–2812.
- Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 1994; 4:39–42.
- Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002; 287:1690–1698.
- Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med 2005; 7:97–104.
- Au N, Rettie AE. Pharmacogenomics of 4-hydroxycoumarin anticoagulants. Drug Metab Rev 2008; 40:355–375.
- García-Martín E, Martínez C, Ladero JM, Agúndez JA. Interethnic and intraethnic variability of CYP2C8 and CYP2C9 polymorphisms in healthy individuals. Mol Diagn Ther 2006; 10:29–40.
- Sullivan-Klose TH, Ghanayem BI, Bell DA, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics 1996; 6:341–349.
- Wen MS, Lee M, Chen JJ, et al. Prospective study of warfarin dosage requirements based on CYP2C9 and VKORC1 genotypes. Clin Pharmacol Ther 2008; 84:83–89.
- Larramendy-Gozalo C, Yang JQ, Verstuyft C, et al. Genetic polymorphism of vitamin K epoxide reductase (VKORC1) 1173C>T in a Chinese and a Caucasian population. Basic Clin Pharmacol Toxicol 2006; 98:611–613.
- Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood 2008; 111:4106–4112.
- Bristol-Myers Squibb. Coumadin (warfarin sodium) Prescribing Information. January 2010.
- Barnes-Jewish Hospital at Washington University Medical Center. Warfarin dosing. http://warfarindosing.orgAccessed 1/20/2011.
- Flockhart DA, O’Kane D, Williams MS, et al. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med 2008; 10:139–150.
- National Institutes of Health. ClinicalTrials.gov. http://clinicaltrials.gov. Accessed January 20, 2011.
- Eckman MH, Rosand J, Greenberg SM, Gage BF. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med 2009; 150:73–83.
- Meckley LM, Gudgeon JM, Anderson JL, Williams MS, Veenstra DL. A policy model to evaluate the benefits, risks, and costs of warfarin pharmacogenomic testing. Pharmacoeconomics 2010; 28:61–74.
- Patrick AR, Avron J, Choudhry NK. Cost-effectiveness of genotype-guided warfarin dosing for patients with atrial fibrillation. Circ Cardiovasc Qual Outcomes 2009; 2:429–436.
- Haas S. New oral Xa and IIa inhibitors: updates on clinical trial results. J Thromb Thrombolysis 2008; 25:52–60.
- Bhatt DL. Tailoring antiplatelet therapy based on pharmacogenomics: how well do the data fit? JAMA 2009; 302:896–897.
- Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009; 302:849–857.
- Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 2009; 373:309–317.
- Simon T, Verstuyft C, Mary-Krause M, et al; French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009; 360:363–375.
- Paré G, Mehta SR, Yusuf S, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med 2010; 363:1704–1714.
- Bristol-Myers Squibb/Sanofi Pharmaceutical Partnership. Plavix (clopidogrel bisulfate) prescribing information. August 2010.
- P450 Drug Interaction Table. Indiana University School of Medicine. http://medicine.iupui.edu/clinpharm/ddis/table.asp. Accessed 1/21/2011.
- Society for Cardiovascular Angiography and Interventions; Holmes DR, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association. Circulation 2010; 122:537–557.
- National Institutes of Health. Genotype Guided Comparison of Clopidogrel and Prasugrel Outcomes Study. http://clinicaltrialsfeeds.org/clinical-trials/show/NCT00995514. Accessed 1/20/2011.
- Amarenco P, Labreuche J. Lipid management in the prevention of stroke: Review and updated meta-analysis of statins for stroke prevention. Lancet Neurol 2009; 8:453–463.
- SEARCH Collaborative Group, Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med 2008; 359:789–799.
- Romaine SP, Bailey KM, Hall AS, Balmforth AJ. The influence of SLC01B1 (OATP1B1) gene polymorphisms on response to statin therapy. Pharmacogenomics J 2010; 10:1–11.
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 2010; Apr 13 [Epub ahead of print].
- Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 2005; 23:9312–9318.
- Beverage JN, Sissung TM, Sion AM, Danesi R, Figg WD. CYP2D6 polymorphisms and the impact on tamoxifen therapy. J Pharm Sci 2007; 96:2224–2231.
- Flockhart D. CYP2D6 genotyping and the pharmacogenetics of tamoxifen. Clin Adv Hematol Oncol 2008; 6:493–494.
- Goetz MP, Kamal A, Ames MM. Tamoxifen pharmacogenomics: the role of CYP2D6 as a predictor of drug response. Clin Pharmacol Ther 2008; 83:160–166.
- Stearns V, Rae JM. Pharmacogenetics and breast cancer endocrine therapy: CYP2D6 as a predictive factor for tamoxifen metabolism and drug response? Expert Rev Mol Med 2008; 10:e34.
- Dezentjé VO, Guchelaar HJ, Nortier JW, van del Velde CJ, Gelderblom H. Clinical implications of CYP2D6 genotyping in tamoxifen treatment for breast cancer. Clin Cancer Res 2009; 15:15–21.
- Higgins MJ, Rae JM, Flockhart DA, Hayes DF, Stearns V. Pharmacogenetics of tamoxifen: who should undergo CYP2D6 genetic testing? J Natl Compr Canc Netw 2009; 7:203–213.
- Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer 2009; 9:576–586.
- Steimer W, Zöpf K, von Amelunxen S, et al. Allele-specific change of concentration and functional gene dose for the prediction of steady-state serum concentrations of amitriptyline and nortriptyline in CYP2C19 and CYP2D6 extensive and intermediate metabolizers. Clinical Cancer 2004; 50:1623–1633.
- Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharm Ther 2008; 83:234–242.
- Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry 2004; 9:442–473.
- Kirchheiner J, Schmidt H, Tzvetkov M, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J 2007; 7:257–265.
- Kirchheiner J. CYP2D6 phenotype prediction from genotype: which system is the best? Clin Pharmacol Ther 2008; 83:225–227.
- Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol 2010; 50:450–458.
- Hoffmann-La Roche Ltd. AmpliChip CYP450 Test. http://www.roche.com/assays/Pages/AmpliChipCYP450Test.aspx. Accessed 1/21/2011.
- Anzenbacher P, Anzenbacherová E. Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci 2001; 58:737–747.
- Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet 2006; 368:704.
- Willmann S, Edginton AN, Coboeken K, Ahr G, Lippert J. Risk to the breast-fed neonate from codeine treatment to the mother: a quantitative mechanistic modeling study. Clin Pharm Ther 2009; 86:634–643.
- Grzesiak M, Beszłej A, Lebioda A, Jonkisz A, Dobosz T, Kienja A. [Retrospective assessment of the antidepressants tolerance in the group of patients with diagnosis of depression and different CYP2D6 genotype.] [In Polish] Psychiatr Pol 2003; 37:433–444.
- Laika B, Leucht S, Heres S, Steimer W. Intermediate metabolizer: increased side effects in psychoactive drug therapy. The key to cost-effectiveness of pretreatment CYP2D6 screening? Pharmacogenomics J 2009; 9:395–403.
- Rau T, Wohlleben G, Wuttke H, et al. CYP2D6 genotype: Impact on adverse effects and nonresponse during treatment with antidepressants—a pilot study. Clin Pharm Ther 2004; 75:386–393.
- McAlpine DE, O’Kane DJ, Black JL, Mrazek DA. Cytochrome P450 2D6 genotype variation and venlafaxine dosage. Mayo Clin Proc 2007; 82:1065–1068.
- Chen S, Chou WH, Blouin RA, et al. The cytochrome P450 2D6 (CYP2D6) enzyme polymorphism: screening costs and influence on clinical outcomes in psychiatry. Clin Pharmacol Ther 1996; 60:522–534.
- Shams ME, Arneth B, Hiemke C, et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther 2006; 31:493–502.
- Whyte EM, Romkes M, Mulsant BH, et al. CYP2D6 genotype and venlafaxine-XR concentrations in depressed elderly. Int J Geriatr Psychiatry 2006; 21:542–549.
- Roberts RL, Mulder RT, Joyce PR, Luty SE, Kennedy MA. No evidence of increased adverse drug reactions in cytochrome P450 CYP2D6 poor metabolizers treated with fluoxetine or nortriptyline. Hum Psychopharmacol 2004; 19:17–23.
- Murphy GM, Kremer C, Rodrigues HE, Schatzberg AF. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry 2003; 160:1830–1835.
- Gillman PK. Re: no evidence of increased adverse drug reactions in cytochrome P450 CYP2D6 poor metabolizers treated with fluoxetine or nortriptyline. Hum Psychopharmacol 2005; 20:61–62.
- Gex-Fabry M, Eap CB, Oneda B, et al. CYP2D6 and ABCB1 genetic variability: influence on paroxetine plasma level and therapeutic response. Ther Drug Monit 2008; 30:474–482.
- Kawanishi C, Lundgren S, Agren H, Bertilsson L. Increased incidence of CYP2D6 gene duplication in patients with persistent mood disorders: ultrarapid metabolism of antidepressants as a cause of nonresponse. A pilot study. Eur J Clin Pharmacol 2004; 59:803–807.
- Serretti A, Calati R, Massat I, et al. Cytochrome P450 CYP1A2, CYP2C9, CYP2C19 and CYP2D6 genes are not associated with response and remission in a sample of depressive patients. Int Clin Psychopharmacol 2009; 24:250–256.
- de Leon J, Armstrong SC, Cozza KL. Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics 2006; 47:75–85.
- de Leon J, Susce MT, Johnson M, et al. DNA microarray technology in the clinical environment: the AmpliChip CYP450 test for CYP2D6 and CYP2C19 genotyping. CNS Spectr 2009; 14:19–34.
- Thuerauf N, Lunkenheimer J. The impact of the CYP2D6-polymorphism on dose recommendations for current antidepressants. Eur Arch Psychiatry Clin Neurosci 2006; 256:287–293.
- Horstmann S, Binder EB. Pharmacogenomics of antidepressant drugs. Pharmacol Ther 2009; 124:57–73.
- Ferreira Ade A, Neves FS, da Rocha FF, et al. The role of 5-HTTLPR polymorphism in antidepressant-associated mania in bipolar disorder. J Affect Disord 2009; 112:267–272.
- Serretti A, Kato M, De Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry 2007; 12:247–257.
- Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry 2010; 15:473–500.
- Reynolds GP, Zhang Z, Zhang X. Polymorphism of the promoter region of the serotonin 5-HT(2C) receptor gene and clozapine-induced weight gain. Am J Psychiatry 2003; 160:677–679.
KEY POINTS
- Polymorphisms that affect the pharmacokinetics and pharmacodynamics of specific drugs are common.
- Testing for certain polymorphisms before prescribing certain drugs could help avoid adverse drug effects and improve efficacy.
- Pharmacogenomic testing has only recently begun to enter clinical practice, and routine testing is currently limited to a few clinical scenarios. However, additional applications may be just around the corner.
- Many pharmacogenomic tests are available, but testing has not yet been recommended for most drugs. Needed are large-scale trials to show that routine testing improves patient outcomes.
Correction: Airway pressure release ventilation
There were several errors in the citation of sources for figures in: Modrykamien A, Chat-burn RL, Ashton RW. Airway pressure release ventilation: An alternative mode of mechanical ventilation in acute respiratory distress syndrome. Cleve Clin J Med 2011; 78:101–110.
For Figure 1, the credit line was omitted. It should be: Reprinted from Papadakos PJ, Lachmann B. The open lung concept of mechanical ventilation: the role of recruitment and stabilization. Crit Care Clin 2007; 23:241–250, with permission from Elsevier.
Figure 2 and Figure 3 were correctly identified as being reprinted from Frawley PM, Habashi NM. Airway pressure release ventilation: theory and practice. AACN Clinical Issues 2001; 12:234–246. However, they were reprinted with permission from Wolters Kluwer Health/Lippincott, Williams & Wilkins, not from Elsevier.
The corrections have been made to the online versions of the article.
There were several errors in the citation of sources for figures in: Modrykamien A, Chat-burn RL, Ashton RW. Airway pressure release ventilation: An alternative mode of mechanical ventilation in acute respiratory distress syndrome. Cleve Clin J Med 2011; 78:101–110.
For Figure 1, the credit line was omitted. It should be: Reprinted from Papadakos PJ, Lachmann B. The open lung concept of mechanical ventilation: the role of recruitment and stabilization. Crit Care Clin 2007; 23:241–250, with permission from Elsevier.
Figure 2 and Figure 3 were correctly identified as being reprinted from Frawley PM, Habashi NM. Airway pressure release ventilation: theory and practice. AACN Clinical Issues 2001; 12:234–246. However, they were reprinted with permission from Wolters Kluwer Health/Lippincott, Williams & Wilkins, not from Elsevier.
The corrections have been made to the online versions of the article.
There were several errors in the citation of sources for figures in: Modrykamien A, Chat-burn RL, Ashton RW. Airway pressure release ventilation: An alternative mode of mechanical ventilation in acute respiratory distress syndrome. Cleve Clin J Med 2011; 78:101–110.
For Figure 1, the credit line was omitted. It should be: Reprinted from Papadakos PJ, Lachmann B. The open lung concept of mechanical ventilation: the role of recruitment and stabilization. Crit Care Clin 2007; 23:241–250, with permission from Elsevier.
Figure 2 and Figure 3 were correctly identified as being reprinted from Frawley PM, Habashi NM. Airway pressure release ventilation: theory and practice. AACN Clinical Issues 2001; 12:234–246. However, they were reprinted with permission from Wolters Kluwer Health/Lippincott, Williams & Wilkins, not from Elsevier.
The corrections have been made to the online versions of the article.
Managing newly diagnosed atrial fibrillation: Rate, rhythm, and risk
Three general concerns dictate the management of atrial fibrillation:
- Controlling the heart rate
- Controlling symptoms
- Preventing thromboembolic events, including stroke.
When seeing a patient with newly diagnosed atrial fibrillation, these same three concerns should be kept in mind, but several additional issues must be addressed:
- Reversible causes of atrial fibrillation must be ruled out
- The true time of onset of the atrial fibrillation and the frequency of the episodes should be ascertained, if possible
- A careful estimation of the patient’s symptom burden should be made.
Atrial fibrillation is common and has a huge impact in terms of the morbidity, death, and costs associated with it. It affects more than 2.2 million Americans.1 Approximately 1 in 10 people over the age of 80 has atrial fibrillation, and for those over the age of 40, the lifetime risk of developing it is one in four.2 Framingham data suggest that the risk of death is approximately twice as high for patients with atrial fibrillation compared with a similar cohort without.3–5
IMPORTANT QUESTIONS DURING THE INITIAL WORKUP
Does the patient have a reversible cause of atrial fibrillation?
Atrial fibrillation is thought to be due to triggers that initiate it and to a myocardial substrate that supports it. While it may develop in the absence of other heart disease, it is often associated with hypertension, diabetes, obesity, structural heart disease (including congenital heart disease), obstructive sleep apnea, advanced age, and alcohol abuse.
Therefore, once atrial fibrillation has been diagnosed, the history, examination, and diagnostic workup should be directed toward looking for potentially reversible causes and for frequently associated comorbidities. Common reversible causes include:
Hyperthyroidism. The laboratory evaluation should include a thyrotropin (thyroid-stimulating hormone, or TSH) level.
Alcohol use, especially binge drinking.
Obstructive sleep apnea, if suspected on the basis of the history or the body habitus.
Structural heart disease such as valvular heart disease or congenital heart defects may also predispose to atrial fibrillation. Therefore, listen carefully to the heart and obtain a transthoracic echocardiogram if one has not already been done or if you suspect a change in valvular disease or systolic function since the most recent study.
How long has the patient been in atrial fibrillation?
The duration of the atrial fibrillation often affects the treatment strategy. Therefore, when the diagnosis has been made, it is important to try to estimate how long the patient has been in atrial fibrillation.
Often, we must settle for an estimate, as the patient’s recollection may be vague. However, in some cases, the symptoms are pronounced or electrocardiographic or telemetric data are available, allowing the time of onset to be clearly defined.
In addition, it is helpful to know if the patient has had prior episodes that were never brought to medical attention. To this end, elicit the patient’s spectrum of symptoms and encourage him or her to think back months or years and try to recall other times when similar symptoms might have occurred.
How do the symptoms affect the patient’s quality of life?
The clinician must also estimate the extent to which the symptoms affect the patient’s quality of life. This is best done when the heart rate is under control. If the patient still has significant symptoms despite adequate rate control, then a rhythm control strategy should probably be pursued.
MANAGING NEWLY DIAGNOSED ATRIAL FIBRILLATION
Control the heart rate with a beta-blocker, a calcium channel blocker, or digoxin
Many patients present during their first episode of atrial fibrillation with a rapid ventricular rate, especially if they are not already taking a drug to slow conduction through the atrioventricular node. If the symptoms are particularly profound, one should try to get the heart rate under control quickly.
Oral agents take time to be absorbed and are not always easy to titrate. Intravenous beta-blockers such as metoprolol (Lopressor) and labetalol (Normodyne, Trandate) or intravenous diltiazem (Cardizem) can slow the heart rate quickly and can be titrated. Once the heart rate is controlled, the oral form can be started, to allow weaning from the intravenous agent. In acute management, we seek a heart rate of less than about 100 to 110 beats per minute.
If the patient’s blood pressure is marginal, loading with intravenous digoxin may be considered. The dosage is 0.5 mg intravenously, then 0.25 mg intravenously in the first 6 hours and another 0.25 mg intravenously in another 6 hours. In patients with renal insufficiency the dosage should be less, or digoxin should be avoided altogether. Often, the blood pressure will improve once the heart rate is decreased, allowing other agents to be initiated. However, if the patient is frankly hypotensive with chest pain, shortness of breath, or a diminished level of consciousness, then emergency electrical cardioversion is indicated even if anticoagulation has not yet been started (more about anticoagulation below).
Oral forms of these same agents are the workhorses for heart rate control in the outpatient setting. Oral beta-blockers and nondihydropyridine calcium channel blockers (ie, diltiazem or verapamil [Calan, Verelan]) are the first-line agents, because when digoxin is used alone, it is relatively poor at controlling the heart rate, especially when the patient is not at rest.
The choice between these agents should be dictated by whether the patient has comorbidities such as coronary artery disease, heart failure, or reactive airway disease. Nondihydropyridine calcium channel blockers are relatively contraindicated in patients with heart failure, while beta-blockers can exacerbate reactive airway disease.6
It is also important to document that the heart rate is adequately controlled outside the hospital or outpatient clinic, where the patient is typically sitting or supine. This can be done with a 6-minute walk, exercise test, or Holter monitor once rate-controlling agents have been titrated.7
When to try to restore sinus rhythm
When atrial fibrillation is first diagnosed, it may not be possible to determine if it is paroxysmal (ie, self-terminating) or persistent. If the episode does not quickly end on its own, consideration may be given to restoring sinus rhythm.
Although experts debate the merits of a rate control approach vs a rhythm control approach for managing atrial fibrillation in the long term, many, including ourselves, recommend trying to restore sinus rhythm at least once when atrial fibrillation is first discovered. It is not always clear if atrial fibrillation is truly asymptomatic. Symptoms such as fatigue or decreased exercise tolerance can be subtle. Additionally, these symptoms may be attributed to other factors such as deconditioning, obesity, or advancing age. Thus, in many cases, only restoring normal sinus rhythm for a time allows the patient and clinician to fully assess the symptoms attributable to atrial fibrillation.
Therefore, in patients with newly diagnosed atrial fibrillation, an attempt to restore sinus rhythm is often warranted. Exceptions are in select patients who have no apparent symptoms and who are very old or are deemed too frail to tolerate cardioversion.
Direct-current cardioversion is typically the treatment of choice when attempting to restore sinus rhythm. The procedure can be done without anticoagulation within 48 hours of the onset of atrial fibrillation, if the time of onset is clear.7 However, clinicians must be careful in defining the onset of atrial fibrillation for this purpose.
Symptoms such as fatigue or shortness of breath can be vague in terms of the exact time of onset and often cannot be relied upon for the purpose of deciding whether cardioversion can be done without anticoagulation. When in doubt, it is best to err on the side of safety and assume that the atrial fibrillation has been going on for more than 48 hours.
If the time of onset is unclear or if more than 48 hours have passed, there are two general strategies for proceeding to electrical cardioversion.
One is to order transesophageal echocardiography and begin anticoagulation therapy at the same time. If there is no thrombus in the left atrium, then cardioversion can be done.8 Therapeutic anticoagulation with heparin, low-molecular-weight heparin, or warfarin (Coumadin) should be achieved within 24 to 48 hours of transesophageal echocardiography and cardioversion to minimize the risk of thromboembolic events, which can occur even after sinus rhythm has been restored.
At our institution, we typically strive to achieve therapeutic anticoagulation with either heparin or low-molecular-weight heparin before cardioversion in this scenario so as to avoid situations in which a patient may undergo cardioversion but then fail to achieve therapeutic anticoagulation for some time due to unforeseen factors.
The other approach is to start warfarin and maintain a goal international normalized ratio (INR) of 2 to 3 for 3 weeks, at which time cardioversion can be performed safely without transesophageal echocardiography.8
Regardless of which strategy is used, anticoagulation should be continued for at least 4 weeks after cardioversion,8 as atrial dysfunction and the risk of stroke may persist for days to weeks after normal sinus rhythm is restored.9
Role of antiarrhythmic drugs
Antiarrhythmic drugs can be used for chemical cardioversion or, more often, to help maintain sinus rhythm after direct-current cardioversion.
Because most of these drugs have at least a small chance of restoring normal sinus rhythm, we need to follow the same rules when starting them as when performing direct-current cardioversion. Patients should not be started on an antiarrhythmic medication until they have had adequate anticoagulation for at least 3 weeks or adequate anticoagulation and a transesophageal echocardiogram confirming that there is no thrombus in the left atrium.
Antiarrhythmic drugs should be started in select patients with newly diagnosed atrial fibrillation in whom a rhythm control strategy will be pursued. For patients whose history suggests a single episode, or episodes that previously self-terminated, an antiarrhythmic may not be necessary. For those with frequent episodes or whose history suggests ongoing atrial fibrillation for a long period, an antiarrhythmic will likely be required to provide a reasonable chance of achieving freedom from atrial fibrillation.
The choice of antiarrhythmic drug should be tailored to the specific patient.
Propafenone (Rythmol) and flecainide (Tambocor) are first-line drugs7 but are contraindicated in patients with coronary artery disease and significant structural heart disease.10
Sotalol (Betapace) and dofetilide (Tikosyn) can be used in patients with coronary artery disease. However, sotalol is contraindicated in patients with congestive heart failure, and dofetilide carries a long list of drug interactions. Both must be used with extreme caution in patients with renal insufficiency, and hospital admission is required for initiation or upward titration of the dose.
Amiodarone (Cordarone) is effective, and in the short term it is typically very well tolerated. However, it has a long half-life, and its potential for long-term toxicity often makes it a poor choice for long-term treatment. The toxicity of amiodarone increases with the cumulative dose. Therefore, this drug should be avoided for long-term therapy of atrial fibrillation in younger patients.
The ‘pill-in-the-pocket’ strategy
The “pill-in-the-pocket” strategy, in which patients are instructed to take their medication only when they have a bout of atrial fibrillation, is a reasonable option for patients with symptomatic recurrences of paroxysmal atrial fibrillation. This strategy is mainly reserved for patients who have relatively infrequent recurrences. Those who have frequent recurrences are usually more effectively treated with daily dosing of an antiarrhythmic. Flecainide and propafenone are the agents of choice for this approach because of their safety profile and efficacy in chemical cardioversion.
While this strategy may be started on an outpatient basis in patients with lone, paroxysmal atrial fibrillation, those with structural heart disease or conduction abnormalities should be observed in the hospital during initiation of therapy to observe for excessive PR prolongation or development of dangerous or worrisome arrhythmias.11–13
Additionally, these agents can decrease the refractory period of the atrioventricular node, thereby increasing the ventricular rate. In the case of atrial flutter, patients may be converted from variable or 2:1 response to a 1:1 conduction. Thus, one should consider also using a beta-blocker with this strategy.
Since the goal of this approach is to convert the patient to sinus rhythm within a few hours of the onset of atrial fibrillation, it may be implemented without the use of warfarin. Patients are instructed that if they do not convert to normal sinus rhythm within a few hours, they should notify the physician so they can undergo electrical cardioversion within the 48-hour window from the onset of atrial fibrillation.
Dronedarone, a new antiarrhythmic drug
Dronedarone (Multaq) is now available and has been shown to be effective in treating atrial fibrillation.14 It has a long half-life and a mechanism of action similar to that of amiodarone. However, it may be inferior to amiodarone in terms of efficacy.15 It is metabolized by CYP3A4. No dosage adjustment is needed for patients with renal insufficiency.
Because dronedarone lacks the iodine moiety found in amiodarone, it should not carry the same toxicity profile. No pulmonary or thyroid toxicity was reported in early trials.16
Nevertheless, dronedarone has several important limitations. It carries a black box warning stating it is contraindicated in patients with severe or recently decompensated heart failure, as the mortality rate was twice as high when this drug was used in such patients in initial studies.17 Additionally, there have been reports of hepatotoxicity requiring liver transplantation in a small number of patients. The extent of this problem and strategies for avoiding it are not yet defined as of the writing of this paper. As with any new medication, patients who are started on dronedarone should be observed closely for any side effects, and these should be reported to assist in the development of the drug’s safety profile.
Pulmonary vein isolation
In a procedure that can potentially cure atrial fibrillation, catheters are inserted into the left atrium and rings of scar tissue are created around the ostia of the pulmonary veins using radiofrequency energy, electrically isolating them from the rest of the left atrium.
Some debate exists as to whether this procedure may be reasonable as a first-line therapy for some patients with atrial fibrillation.18,19 It may be considered as an early treatment strategy in a small subset of patients, specifically young patients with symptomatic, recurrent atrial fibrillation, especially if they are averse to long-term antiarrhythmic therapy.
Because patients may still be more prone to atrial arrhythmias for several weeks to months after the procedure, they must be able to tolerate anticoagulation with warfarin for at least several months.
Rate control vs rhythm control
The choice between a rate control strategy or a rhythm control strategy in the long term is not always straightforward. While atrial fibrillation is clearly associated with higher morbidity and mortality rates, there are few data to date showing that restoring and maintaining sinus rhythm in patients with atrial fibrillation reduce the incidence of morbid complications or the likelihood of death.
Thus, current guidelines recommend a rate control strategy in patients who have no symptoms, and a rhythm control strategy if rate control cannot be achieved or if symptoms persist despite adequate control of the heart rate.7 The circumstances and preferences of the individual patient should carry weight in this decision.
Trials are under way that may shed more light on the relative benefits of rhythm control with ablation or antiarrhythmics and rate control.
PREVENTING THROMBOEMBOLIC EVENTS
Warfarin
In the short term, warfarin therapy may be dictated by plans to restore sinus rhythm. Patients need warfarin for at least 4 weeks after cardioversion unless it is performed within 48 hours of the onset of atrial fibrillation.
The CHADS2score (1 point each for congestive heart failure, hypertension, age 75 or older, and diabetes mellitus; 2 points for prior stroke or transient ischemic attack) is useful when deciding whether to give long-term anticoagulation.
For patients with a score of 0, the risk of stroke is lower than the risk of a major bleeding complication while on therapeutic warfarin.20,21 For these patients, aspirin 81 to 325 mg daily is recommended for stroke prophylaxis.
For those with a score of 2 or greater, the risk of stroke without warfarin is greater than the risk of a major bleeding complication with warfarin. These patients should receive warfarin with a goal INR of 2.0 to 3.0.7
Patients with a CHADS2 score of 1 present a dilemma, as their risk of stroke without warfarin is about the same as their risk of a major bleeding complication with warfarin. They can be managed with either warfarin or aspirin, according to the physician’s judgment.7 In these cases, factors such as hobbies or professions that might increase the risk of bleeding, perceived frequency of atrial fibrillation episodes, and even patient preconceptions about warfarin are often used when deciding between aspirin and warfarin.
Patients with a CHADS2 score of 2 or greater with a single episode of atrial fibrillation and a likely reversible cause may also pose a dilemma when deciding whether to start warfarin. These patients have demonstrated they at least have the substrate to maintain atrial fibrillation. This situation again calls for physician judgment. Bear in mind that asymptomatic recurrences are common in patients with atrial fibrillation.22,23 A higher CHADS2 score denotes a greater risk of stroke and may influence this decision. It is usually beneficial to enlist the patient in this decision-making process, as patients often have very strong opinions about tolerance of the risk of stroke or of warfarin therapy itself.
Another strategy is to start anticoagulation with warfarin and aggressively monitor for recurrences. If the patient has no recurrences of atrial fibrillation after 6 to 12 months and the reversible cause is resolved, one can then revisit the need for warfarin.
Role of aspirin and clopidogrel
Aspirin, alone or in conjunction with clopidogrel (Plavix), may provide an alternative for stroke prophylaxis in patients in whom warfarin is contraindicated. While inferior to warfarin, the combination of aspirin and clopidogrel has been shown to decrease the incidence of major thromboembolic events, especially stroke.24 However, the risk of a major bleeding complication was also significantly increased.
This combination may be a reasonable strategy in select patients with a CHADS2 score of 2 or greater in whom warfarin cannot be used for reasons such as personal aversion to the medication, side effects, or nonbleeding complications or in patients whose INR is exceedingly difficult to keep within the therapeutic range.
Dabigatran, a new anticoagulant
The newest option for anticoagulation in patients with atrial fibrillation is a direct thrombin inhibitor, dabigatran (Pradaxa).
In the Randomized Evaluation of Long-Term Anticoagulation Therapy (RE-LY) trial,25 dabigatran was studied head-to-head with warfarin. The doses of dabigatran studied were 110 mg and 150 mg twice a day. At 150 mg twice a day, patients on dabigatran had a lower rate of stroke than with warfarin (1.11% vs 1.69%, P < .001), as well as a lower rate of central nervous system bleeding (0.10% vs 0.38% with warfarin, P < .001). The rates of major bleeding were comparable in the patients receiving warfarin or dabigatran 150 mg twice a day, but the rate of gastrointestinal bleeding was higher in the dabigatran group (1.51% vs 1.02% with warfarin, P < .001).25
Dabigatran was recently approved by the US Food and Drug Administration for use in patients with atrial fibrillation. Doses of 150 mg and 75 mg are available.
Dabigatran is renally excreted, and the 150 mg twice-a-day dosing is intended for patients with a creatinine clearance greater than 30 mL/min. The 75-mg twice-a-day dosing is intended for patients with a creatinine clearance of 15 to 30 mL/min. However, it should be noted that currently there are no data to support the 75-mg twice-a-day dosing.
Dabigatran does have several advantages over warfarin. Patients do not need to avoid foods containing vitamin K, and routine serial monitoring does not appear to be needed. As with any new medication, patients who are started on dabigatran should be observed closely for any side effects, and these should be reported to assist in the development of the drug’s safety profile.
SPECIAL CIRCUMSTANCES
After cardiac or noncardiac surgery
Atrial fibrillation is common after open heart surgery, occurring in approximately 25% to 50% of patients.26–28
When this happens, at least one or two attempts are made to restore sinus rhythm. Especially in the early postoperative period, anticoagulation with heparin or warfarin may be contraindicated, so careful attention must be paid to the patient’s heart rhythm so that atrial fibrillation can be recognized quickly and cardioversion performed within a 48-hour window of onset. Beta-blockers, diltiazem, and verapamil are typically used for rate control.
When atrial fibrillation recurs in patients who have undergone open heart surgery, antiarrhythmics are started early to help prevent further recurrences. At our institution, we usually use amiodarone, as it is highly effective and well tolerated in the short term. When started on amiodarone for postoperative atrial fibrillation, patients are informed that the drug will be stopped after about 2 to 3 months. For patients who continue to have bouts of atrial fibrillation, the need for antiarrhythmic medications can be reassessed, and, if needed, the optimal antiarrhythmic medication for long-term therapy for the patient can be chosen.
Atrial fibrillation in severe, acute illness
Atrial fibrillation is common in the setting of extreme systemic stressors such as shock and sepsis and when the patient is being supported with inotropic agents. In this setting, patients may be in a high-catecholamine state, and both the heart rate and the heart rhythm may be very difficult to control.
Beta-blockers and nondihydropyridine calcium channel blockers should not be used when patients are on medications to support blood pressure, and in this setting, when the patient’s hemodynamic status permits the use of these agents, their effect may be minimal.
Amiodarone or perhaps digoxin may slow the heart rate somewhat without too much effect on the blood pressure. However, with amiodarone, one may have to accept a risk of chemical cardioversion.
Electrical cardioversion with or without the assistance of an antiarrhythmic drug may control the heart rate by restoring sinus rhythm. However, atrial fibrillation often recurs, and if it recurs quickly one may have to accept elevated heart rates until the underlying process is addressed.
- Go AS, Hylek EM, Phillips KA, et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the Anticoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA 2001; 285:2370–2375.
- Lloyd-Jones DM, Wang TJ, Leip EP, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation 2004; 110:1042–1046.
- Benjamin EJ, Wolf PA, D’Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation 1998; 98:946–952.
- Kannel WB, Wolf PA, Benjamin EJ, Levy D. Prevalence, incidence, prognosis, and predisposing conditions for atrial fibrillation: population-based estimates. Am J Cardiol 1998; 82( 8A):2N–9N.
- Kannel WB, Abbott RD, Savage DD, McNamara PM. Epidemiologic features of chronic atrial fibrillation: the Framingham study. N Engl J Med 1982; 306:1018–1022.
- The Multicenter Diltiazem Postinfarction Trial Research Group. The effect of diltiazem on mortality and reinfarction after myocardial infarction. N Engl J Med 1988; 319:385–392.
- European heart Rhythm Association; Heart Rhythm society, Fuster V, Rydén LE, Cannom DS, et al. ACC/AHA/ESC guidelines for the management of patients with atrial fibrillation—executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation). J Am Coll Cardiol 2006; 48:854–906.
- Klein AL, Grimm RA, Murray RD, et al; Assessment of Cardioversion Using Transesophageal Echocardiography Investigators. Use of transesophageal echocardiography to guide cardioversion in patients with atrial fibrillation. N Engl J Med 2001; 344:1411–1120.
- Grimm RA, Leung DY, Black IW, Stewart WJ, Thomas JD, Klein AL. Left atrial appendage “stunning” after spontaneous conversion of atrial fibrillation demonstrated by transesophageal Doppler echocardiography. Am Heart J 1995; 130:174–176.
- The Cardiac Arrhythmia Suppression Trial (CAST) Investigators. Preliminary report: effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. N Engl J Med 1989; 321:406–412.
- Alboni P, Tomasi C, Menozzi C, et al. Efficacy and safety of out-of-hospital self-administered single-dose oral drug treatment in the management of infrequent, well-tolerated paroxysmal supraventricular tachycardia. J Am Coll Cardiol 2001; 37:548–553.
- Capucci A, Villani GQ, Piepoli MF. Reproducible efficacy of loading oral propafenone in restoring sinus rhythm in patients with paroxysmal atrial fibrillation. Am J Cardiol 2003; 92:1345–1347.
- Khan IA. Single oral loading dose of propafenone for pharmacological cardioversion of recent-onset atrial fibrillation. J Am Coll Cardiol 2001; 37:542–547.
- Singh BN, Connolly SJ, Crijns HJ, et al; EURIDIS and ADONIS Investigators. Dronedarone for maintenance of sinus rhythm in atrial fibrillation or flutter. N Engl J Med 2007; 357:987–999.
- Le Heuzey J, De Ferrari GM, Radzik D, Santini M, Zhu J, Davy JM. A short-term, randomized, double-blind, parallel-group study to evaluate the efficacy and safety of dronedarone versus amiodarone in patients with persistent atrial fibrillation: the DIONYSOS study. J Cardiovasc Electrophysiol 2010; 21:597–605.
- Hohnloser SH, Crijns HJ, van Eickels M, et al. Effect of dronedarone on cardiovascular events in atrial fibrillation. N Engl J Med 2009; 360:668–678.
- Køber L, Torp-Pedersen C, McMurray JJ, et al; Dronedarone Study Group. Increased mortality after dronedarone therapy for severe heart failure. N Engl J Med 2008; 358:2678–2687.
- Pappone C, Rosanio S, Augello G, et al. Mortality, morbidity, and quality of life after circumferential pulmonary vein ablation for atrial fibrillation: outcomes from a controlled nonrandomized long-term study. J Am Coll Cardiol 2003; 42:185–197.
- Wazni OM, Marrouche NF, Martin DO, et al. Radiofrequency ablation vs antiarrhythmic drugs as first-line treatment of symptomatic atrial fibrillation: a randomized trial. JAMA 2005; 293:2634–2640.
- van Walraven C, Hart RG, Singer DE, et al. Oral anticoagulants vs aspirin in nonvalvular atrial fibrillation: an individual patient metaanalysis. JAMA 2002; 288:2441–2448.
- Hart RG, Benavente O, McBride R, Pearce LA. Antithrombotic therapy to prevent stroke in patients with atrial fibrillation: a metaanalysis. Ann Intern Med 1999; 131:492–501.
- Page RL, Wilkinson WE, Clair WK, McCarthy EA, Pritchett EL. Asymptomatic arrhythmias in patients with symptomatic paroxysmal atrial fibrillation and paroxysmal supraventricular tachycardia. Circulation 1994; 89:224–227.
- Savelieva I, Camm AJ. Clinical relevance of silent atrial fibrillation: prevalence, prognosis, quality of life, and management. J Intervent Card Electrophysiol 2000; 4:369–382.
- ACTIVE Investigators, Connolly SJ, Pogue J, Hart RG, et al. Effect of clopidogrel added to aspirin in patients with atrial fibrillation. N Engl J Med 2009; 360:2066–2078.
- Connolly SJ, Ezekowitz MD, Yusuf S, et al; RE-LY Steering Committee and Investigators. Dabigatran versus warfarin in patients with atrial fibrillation. N Engl J Med 2009; 361:1139–1151. Erratum in: N Engl J Med 2010; 363:1877.
- Almassi GH, Schowalter T, Nicolosi AC, et al. Atrial fibrillation after cardiac surgery: a major morbid event? Ann Surg 1997; 226:501–511.
- Creswell LL, Schuessler RB, Rosenbloom M, Cox JL. Hazards of postoperative atrial arrhythmias. Ann Thorac Surg 1993; 56:539–549.
- Mathew JP, Fontes ML, Tudor IC, et al; Investigators of the Ischemia Research and Education Foundation; Multicenter Study of Perioperative Ischemia Research Group. A multicenter risk index for atrial fibrillation after cardiac surgery. JAMA 2004; 291:1720–1729.
Three general concerns dictate the management of atrial fibrillation:
- Controlling the heart rate
- Controlling symptoms
- Preventing thromboembolic events, including stroke.
When seeing a patient with newly diagnosed atrial fibrillation, these same three concerns should be kept in mind, but several additional issues must be addressed:
- Reversible causes of atrial fibrillation must be ruled out
- The true time of onset of the atrial fibrillation and the frequency of the episodes should be ascertained, if possible
- A careful estimation of the patient’s symptom burden should be made.
Atrial fibrillation is common and has a huge impact in terms of the morbidity, death, and costs associated with it. It affects more than 2.2 million Americans.1 Approximately 1 in 10 people over the age of 80 has atrial fibrillation, and for those over the age of 40, the lifetime risk of developing it is one in four.2 Framingham data suggest that the risk of death is approximately twice as high for patients with atrial fibrillation compared with a similar cohort without.3–5
IMPORTANT QUESTIONS DURING THE INITIAL WORKUP
Does the patient have a reversible cause of atrial fibrillation?
Atrial fibrillation is thought to be due to triggers that initiate it and to a myocardial substrate that supports it. While it may develop in the absence of other heart disease, it is often associated with hypertension, diabetes, obesity, structural heart disease (including congenital heart disease), obstructive sleep apnea, advanced age, and alcohol abuse.
Therefore, once atrial fibrillation has been diagnosed, the history, examination, and diagnostic workup should be directed toward looking for potentially reversible causes and for frequently associated comorbidities. Common reversible causes include:
Hyperthyroidism. The laboratory evaluation should include a thyrotropin (thyroid-stimulating hormone, or TSH) level.
Alcohol use, especially binge drinking.
Obstructive sleep apnea, if suspected on the basis of the history or the body habitus.
Structural heart disease such as valvular heart disease or congenital heart defects may also predispose to atrial fibrillation. Therefore, listen carefully to the heart and obtain a transthoracic echocardiogram if one has not already been done or if you suspect a change in valvular disease or systolic function since the most recent study.
How long has the patient been in atrial fibrillation?
The duration of the atrial fibrillation often affects the treatment strategy. Therefore, when the diagnosis has been made, it is important to try to estimate how long the patient has been in atrial fibrillation.
Often, we must settle for an estimate, as the patient’s recollection may be vague. However, in some cases, the symptoms are pronounced or electrocardiographic or telemetric data are available, allowing the time of onset to be clearly defined.
In addition, it is helpful to know if the patient has had prior episodes that were never brought to medical attention. To this end, elicit the patient’s spectrum of symptoms and encourage him or her to think back months or years and try to recall other times when similar symptoms might have occurred.
How do the symptoms affect the patient’s quality of life?
The clinician must also estimate the extent to which the symptoms affect the patient’s quality of life. This is best done when the heart rate is under control. If the patient still has significant symptoms despite adequate rate control, then a rhythm control strategy should probably be pursued.
MANAGING NEWLY DIAGNOSED ATRIAL FIBRILLATION
Control the heart rate with a beta-blocker, a calcium channel blocker, or digoxin
Many patients present during their first episode of atrial fibrillation with a rapid ventricular rate, especially if they are not already taking a drug to slow conduction through the atrioventricular node. If the symptoms are particularly profound, one should try to get the heart rate under control quickly.
Oral agents take time to be absorbed and are not always easy to titrate. Intravenous beta-blockers such as metoprolol (Lopressor) and labetalol (Normodyne, Trandate) or intravenous diltiazem (Cardizem) can slow the heart rate quickly and can be titrated. Once the heart rate is controlled, the oral form can be started, to allow weaning from the intravenous agent. In acute management, we seek a heart rate of less than about 100 to 110 beats per minute.
If the patient’s blood pressure is marginal, loading with intravenous digoxin may be considered. The dosage is 0.5 mg intravenously, then 0.25 mg intravenously in the first 6 hours and another 0.25 mg intravenously in another 6 hours. In patients with renal insufficiency the dosage should be less, or digoxin should be avoided altogether. Often, the blood pressure will improve once the heart rate is decreased, allowing other agents to be initiated. However, if the patient is frankly hypotensive with chest pain, shortness of breath, or a diminished level of consciousness, then emergency electrical cardioversion is indicated even if anticoagulation has not yet been started (more about anticoagulation below).
Oral forms of these same agents are the workhorses for heart rate control in the outpatient setting. Oral beta-blockers and nondihydropyridine calcium channel blockers (ie, diltiazem or verapamil [Calan, Verelan]) are the first-line agents, because when digoxin is used alone, it is relatively poor at controlling the heart rate, especially when the patient is not at rest.
The choice between these agents should be dictated by whether the patient has comorbidities such as coronary artery disease, heart failure, or reactive airway disease. Nondihydropyridine calcium channel blockers are relatively contraindicated in patients with heart failure, while beta-blockers can exacerbate reactive airway disease.6
It is also important to document that the heart rate is adequately controlled outside the hospital or outpatient clinic, where the patient is typically sitting or supine. This can be done with a 6-minute walk, exercise test, or Holter monitor once rate-controlling agents have been titrated.7
When to try to restore sinus rhythm
When atrial fibrillation is first diagnosed, it may not be possible to determine if it is paroxysmal (ie, self-terminating) or persistent. If the episode does not quickly end on its own, consideration may be given to restoring sinus rhythm.
Although experts debate the merits of a rate control approach vs a rhythm control approach for managing atrial fibrillation in the long term, many, including ourselves, recommend trying to restore sinus rhythm at least once when atrial fibrillation is first discovered. It is not always clear if atrial fibrillation is truly asymptomatic. Symptoms such as fatigue or decreased exercise tolerance can be subtle. Additionally, these symptoms may be attributed to other factors such as deconditioning, obesity, or advancing age. Thus, in many cases, only restoring normal sinus rhythm for a time allows the patient and clinician to fully assess the symptoms attributable to atrial fibrillation.
Therefore, in patients with newly diagnosed atrial fibrillation, an attempt to restore sinus rhythm is often warranted. Exceptions are in select patients who have no apparent symptoms and who are very old or are deemed too frail to tolerate cardioversion.
Direct-current cardioversion is typically the treatment of choice when attempting to restore sinus rhythm. The procedure can be done without anticoagulation within 48 hours of the onset of atrial fibrillation, if the time of onset is clear.7 However, clinicians must be careful in defining the onset of atrial fibrillation for this purpose.
Symptoms such as fatigue or shortness of breath can be vague in terms of the exact time of onset and often cannot be relied upon for the purpose of deciding whether cardioversion can be done without anticoagulation. When in doubt, it is best to err on the side of safety and assume that the atrial fibrillation has been going on for more than 48 hours.
If the time of onset is unclear or if more than 48 hours have passed, there are two general strategies for proceeding to electrical cardioversion.
One is to order transesophageal echocardiography and begin anticoagulation therapy at the same time. If there is no thrombus in the left atrium, then cardioversion can be done.8 Therapeutic anticoagulation with heparin, low-molecular-weight heparin, or warfarin (Coumadin) should be achieved within 24 to 48 hours of transesophageal echocardiography and cardioversion to minimize the risk of thromboembolic events, which can occur even after sinus rhythm has been restored.
At our institution, we typically strive to achieve therapeutic anticoagulation with either heparin or low-molecular-weight heparin before cardioversion in this scenario so as to avoid situations in which a patient may undergo cardioversion but then fail to achieve therapeutic anticoagulation for some time due to unforeseen factors.
The other approach is to start warfarin and maintain a goal international normalized ratio (INR) of 2 to 3 for 3 weeks, at which time cardioversion can be performed safely without transesophageal echocardiography.8
Regardless of which strategy is used, anticoagulation should be continued for at least 4 weeks after cardioversion,8 as atrial dysfunction and the risk of stroke may persist for days to weeks after normal sinus rhythm is restored.9
Role of antiarrhythmic drugs
Antiarrhythmic drugs can be used for chemical cardioversion or, more often, to help maintain sinus rhythm after direct-current cardioversion.
Because most of these drugs have at least a small chance of restoring normal sinus rhythm, we need to follow the same rules when starting them as when performing direct-current cardioversion. Patients should not be started on an antiarrhythmic medication until they have had adequate anticoagulation for at least 3 weeks or adequate anticoagulation and a transesophageal echocardiogram confirming that there is no thrombus in the left atrium.
Antiarrhythmic drugs should be started in select patients with newly diagnosed atrial fibrillation in whom a rhythm control strategy will be pursued. For patients whose history suggests a single episode, or episodes that previously self-terminated, an antiarrhythmic may not be necessary. For those with frequent episodes or whose history suggests ongoing atrial fibrillation for a long period, an antiarrhythmic will likely be required to provide a reasonable chance of achieving freedom from atrial fibrillation.
The choice of antiarrhythmic drug should be tailored to the specific patient.
Propafenone (Rythmol) and flecainide (Tambocor) are first-line drugs7 but are contraindicated in patients with coronary artery disease and significant structural heart disease.10
Sotalol (Betapace) and dofetilide (Tikosyn) can be used in patients with coronary artery disease. However, sotalol is contraindicated in patients with congestive heart failure, and dofetilide carries a long list of drug interactions. Both must be used with extreme caution in patients with renal insufficiency, and hospital admission is required for initiation or upward titration of the dose.
Amiodarone (Cordarone) is effective, and in the short term it is typically very well tolerated. However, it has a long half-life, and its potential for long-term toxicity often makes it a poor choice for long-term treatment. The toxicity of amiodarone increases with the cumulative dose. Therefore, this drug should be avoided for long-term therapy of atrial fibrillation in younger patients.
The ‘pill-in-the-pocket’ strategy
The “pill-in-the-pocket” strategy, in which patients are instructed to take their medication only when they have a bout of atrial fibrillation, is a reasonable option for patients with symptomatic recurrences of paroxysmal atrial fibrillation. This strategy is mainly reserved for patients who have relatively infrequent recurrences. Those who have frequent recurrences are usually more effectively treated with daily dosing of an antiarrhythmic. Flecainide and propafenone are the agents of choice for this approach because of their safety profile and efficacy in chemical cardioversion.
While this strategy may be started on an outpatient basis in patients with lone, paroxysmal atrial fibrillation, those with structural heart disease or conduction abnormalities should be observed in the hospital during initiation of therapy to observe for excessive PR prolongation or development of dangerous or worrisome arrhythmias.11–13
Additionally, these agents can decrease the refractory period of the atrioventricular node, thereby increasing the ventricular rate. In the case of atrial flutter, patients may be converted from variable or 2:1 response to a 1:1 conduction. Thus, one should consider also using a beta-blocker with this strategy.
Since the goal of this approach is to convert the patient to sinus rhythm within a few hours of the onset of atrial fibrillation, it may be implemented without the use of warfarin. Patients are instructed that if they do not convert to normal sinus rhythm within a few hours, they should notify the physician so they can undergo electrical cardioversion within the 48-hour window from the onset of atrial fibrillation.
Dronedarone, a new antiarrhythmic drug
Dronedarone (Multaq) is now available and has been shown to be effective in treating atrial fibrillation.14 It has a long half-life and a mechanism of action similar to that of amiodarone. However, it may be inferior to amiodarone in terms of efficacy.15 It is metabolized by CYP3A4. No dosage adjustment is needed for patients with renal insufficiency.
Because dronedarone lacks the iodine moiety found in amiodarone, it should not carry the same toxicity profile. No pulmonary or thyroid toxicity was reported in early trials.16
Nevertheless, dronedarone has several important limitations. It carries a black box warning stating it is contraindicated in patients with severe or recently decompensated heart failure, as the mortality rate was twice as high when this drug was used in such patients in initial studies.17 Additionally, there have been reports of hepatotoxicity requiring liver transplantation in a small number of patients. The extent of this problem and strategies for avoiding it are not yet defined as of the writing of this paper. As with any new medication, patients who are started on dronedarone should be observed closely for any side effects, and these should be reported to assist in the development of the drug’s safety profile.
Pulmonary vein isolation
In a procedure that can potentially cure atrial fibrillation, catheters are inserted into the left atrium and rings of scar tissue are created around the ostia of the pulmonary veins using radiofrequency energy, electrically isolating them from the rest of the left atrium.
Some debate exists as to whether this procedure may be reasonable as a first-line therapy for some patients with atrial fibrillation.18,19 It may be considered as an early treatment strategy in a small subset of patients, specifically young patients with symptomatic, recurrent atrial fibrillation, especially if they are averse to long-term antiarrhythmic therapy.
Because patients may still be more prone to atrial arrhythmias for several weeks to months after the procedure, they must be able to tolerate anticoagulation with warfarin for at least several months.
Rate control vs rhythm control
The choice between a rate control strategy or a rhythm control strategy in the long term is not always straightforward. While atrial fibrillation is clearly associated with higher morbidity and mortality rates, there are few data to date showing that restoring and maintaining sinus rhythm in patients with atrial fibrillation reduce the incidence of morbid complications or the likelihood of death.
Thus, current guidelines recommend a rate control strategy in patients who have no symptoms, and a rhythm control strategy if rate control cannot be achieved or if symptoms persist despite adequate control of the heart rate.7 The circumstances and preferences of the individual patient should carry weight in this decision.
Trials are under way that may shed more light on the relative benefits of rhythm control with ablation or antiarrhythmics and rate control.
PREVENTING THROMBOEMBOLIC EVENTS
Warfarin
In the short term, warfarin therapy may be dictated by plans to restore sinus rhythm. Patients need warfarin for at least 4 weeks after cardioversion unless it is performed within 48 hours of the onset of atrial fibrillation.
The CHADS2score (1 point each for congestive heart failure, hypertension, age 75 or older, and diabetes mellitus; 2 points for prior stroke or transient ischemic attack) is useful when deciding whether to give long-term anticoagulation.
For patients with a score of 0, the risk of stroke is lower than the risk of a major bleeding complication while on therapeutic warfarin.20,21 For these patients, aspirin 81 to 325 mg daily is recommended for stroke prophylaxis.
For those with a score of 2 or greater, the risk of stroke without warfarin is greater than the risk of a major bleeding complication with warfarin. These patients should receive warfarin with a goal INR of 2.0 to 3.0.7
Patients with a CHADS2 score of 1 present a dilemma, as their risk of stroke without warfarin is about the same as their risk of a major bleeding complication with warfarin. They can be managed with either warfarin or aspirin, according to the physician’s judgment.7 In these cases, factors such as hobbies or professions that might increase the risk of bleeding, perceived frequency of atrial fibrillation episodes, and even patient preconceptions about warfarin are often used when deciding between aspirin and warfarin.
Patients with a CHADS2 score of 2 or greater with a single episode of atrial fibrillation and a likely reversible cause may also pose a dilemma when deciding whether to start warfarin. These patients have demonstrated they at least have the substrate to maintain atrial fibrillation. This situation again calls for physician judgment. Bear in mind that asymptomatic recurrences are common in patients with atrial fibrillation.22,23 A higher CHADS2 score denotes a greater risk of stroke and may influence this decision. It is usually beneficial to enlist the patient in this decision-making process, as patients often have very strong opinions about tolerance of the risk of stroke or of warfarin therapy itself.
Another strategy is to start anticoagulation with warfarin and aggressively monitor for recurrences. If the patient has no recurrences of atrial fibrillation after 6 to 12 months and the reversible cause is resolved, one can then revisit the need for warfarin.
Role of aspirin and clopidogrel
Aspirin, alone or in conjunction with clopidogrel (Plavix), may provide an alternative for stroke prophylaxis in patients in whom warfarin is contraindicated. While inferior to warfarin, the combination of aspirin and clopidogrel has been shown to decrease the incidence of major thromboembolic events, especially stroke.24 However, the risk of a major bleeding complication was also significantly increased.
This combination may be a reasonable strategy in select patients with a CHADS2 score of 2 or greater in whom warfarin cannot be used for reasons such as personal aversion to the medication, side effects, or nonbleeding complications or in patients whose INR is exceedingly difficult to keep within the therapeutic range.
Dabigatran, a new anticoagulant
The newest option for anticoagulation in patients with atrial fibrillation is a direct thrombin inhibitor, dabigatran (Pradaxa).
In the Randomized Evaluation of Long-Term Anticoagulation Therapy (RE-LY) trial,25 dabigatran was studied head-to-head with warfarin. The doses of dabigatran studied were 110 mg and 150 mg twice a day. At 150 mg twice a day, patients on dabigatran had a lower rate of stroke than with warfarin (1.11% vs 1.69%, P < .001), as well as a lower rate of central nervous system bleeding (0.10% vs 0.38% with warfarin, P < .001). The rates of major bleeding were comparable in the patients receiving warfarin or dabigatran 150 mg twice a day, but the rate of gastrointestinal bleeding was higher in the dabigatran group (1.51% vs 1.02% with warfarin, P < .001).25
Dabigatran was recently approved by the US Food and Drug Administration for use in patients with atrial fibrillation. Doses of 150 mg and 75 mg are available.
Dabigatran is renally excreted, and the 150 mg twice-a-day dosing is intended for patients with a creatinine clearance greater than 30 mL/min. The 75-mg twice-a-day dosing is intended for patients with a creatinine clearance of 15 to 30 mL/min. However, it should be noted that currently there are no data to support the 75-mg twice-a-day dosing.
Dabigatran does have several advantages over warfarin. Patients do not need to avoid foods containing vitamin K, and routine serial monitoring does not appear to be needed. As with any new medication, patients who are started on dabigatran should be observed closely for any side effects, and these should be reported to assist in the development of the drug’s safety profile.
SPECIAL CIRCUMSTANCES
After cardiac or noncardiac surgery
Atrial fibrillation is common after open heart surgery, occurring in approximately 25% to 50% of patients.26–28
When this happens, at least one or two attempts are made to restore sinus rhythm. Especially in the early postoperative period, anticoagulation with heparin or warfarin may be contraindicated, so careful attention must be paid to the patient’s heart rhythm so that atrial fibrillation can be recognized quickly and cardioversion performed within a 48-hour window of onset. Beta-blockers, diltiazem, and verapamil are typically used for rate control.
When atrial fibrillation recurs in patients who have undergone open heart surgery, antiarrhythmics are started early to help prevent further recurrences. At our institution, we usually use amiodarone, as it is highly effective and well tolerated in the short term. When started on amiodarone for postoperative atrial fibrillation, patients are informed that the drug will be stopped after about 2 to 3 months. For patients who continue to have bouts of atrial fibrillation, the need for antiarrhythmic medications can be reassessed, and, if needed, the optimal antiarrhythmic medication for long-term therapy for the patient can be chosen.
Atrial fibrillation in severe, acute illness
Atrial fibrillation is common in the setting of extreme systemic stressors such as shock and sepsis and when the patient is being supported with inotropic agents. In this setting, patients may be in a high-catecholamine state, and both the heart rate and the heart rhythm may be very difficult to control.
Beta-blockers and nondihydropyridine calcium channel blockers should not be used when patients are on medications to support blood pressure, and in this setting, when the patient’s hemodynamic status permits the use of these agents, their effect may be minimal.
Amiodarone or perhaps digoxin may slow the heart rate somewhat without too much effect on the blood pressure. However, with amiodarone, one may have to accept a risk of chemical cardioversion.
Electrical cardioversion with or without the assistance of an antiarrhythmic drug may control the heart rate by restoring sinus rhythm. However, atrial fibrillation often recurs, and if it recurs quickly one may have to accept elevated heart rates until the underlying process is addressed.
Three general concerns dictate the management of atrial fibrillation:
- Controlling the heart rate
- Controlling symptoms
- Preventing thromboembolic events, including stroke.
When seeing a patient with newly diagnosed atrial fibrillation, these same three concerns should be kept in mind, but several additional issues must be addressed:
- Reversible causes of atrial fibrillation must be ruled out
- The true time of onset of the atrial fibrillation and the frequency of the episodes should be ascertained, if possible
- A careful estimation of the patient’s symptom burden should be made.
Atrial fibrillation is common and has a huge impact in terms of the morbidity, death, and costs associated with it. It affects more than 2.2 million Americans.1 Approximately 1 in 10 people over the age of 80 has atrial fibrillation, and for those over the age of 40, the lifetime risk of developing it is one in four.2 Framingham data suggest that the risk of death is approximately twice as high for patients with atrial fibrillation compared with a similar cohort without.3–5
IMPORTANT QUESTIONS DURING THE INITIAL WORKUP
Does the patient have a reversible cause of atrial fibrillation?
Atrial fibrillation is thought to be due to triggers that initiate it and to a myocardial substrate that supports it. While it may develop in the absence of other heart disease, it is often associated with hypertension, diabetes, obesity, structural heart disease (including congenital heart disease), obstructive sleep apnea, advanced age, and alcohol abuse.
Therefore, once atrial fibrillation has been diagnosed, the history, examination, and diagnostic workup should be directed toward looking for potentially reversible causes and for frequently associated comorbidities. Common reversible causes include:
Hyperthyroidism. The laboratory evaluation should include a thyrotropin (thyroid-stimulating hormone, or TSH) level.
Alcohol use, especially binge drinking.
Obstructive sleep apnea, if suspected on the basis of the history or the body habitus.
Structural heart disease such as valvular heart disease or congenital heart defects may also predispose to atrial fibrillation. Therefore, listen carefully to the heart and obtain a transthoracic echocardiogram if one has not already been done or if you suspect a change in valvular disease or systolic function since the most recent study.
How long has the patient been in atrial fibrillation?
The duration of the atrial fibrillation often affects the treatment strategy. Therefore, when the diagnosis has been made, it is important to try to estimate how long the patient has been in atrial fibrillation.
Often, we must settle for an estimate, as the patient’s recollection may be vague. However, in some cases, the symptoms are pronounced or electrocardiographic or telemetric data are available, allowing the time of onset to be clearly defined.
In addition, it is helpful to know if the patient has had prior episodes that were never brought to medical attention. To this end, elicit the patient’s spectrum of symptoms and encourage him or her to think back months or years and try to recall other times when similar symptoms might have occurred.
How do the symptoms affect the patient’s quality of life?
The clinician must also estimate the extent to which the symptoms affect the patient’s quality of life. This is best done when the heart rate is under control. If the patient still has significant symptoms despite adequate rate control, then a rhythm control strategy should probably be pursued.
MANAGING NEWLY DIAGNOSED ATRIAL FIBRILLATION
Control the heart rate with a beta-blocker, a calcium channel blocker, or digoxin
Many patients present during their first episode of atrial fibrillation with a rapid ventricular rate, especially if they are not already taking a drug to slow conduction through the atrioventricular node. If the symptoms are particularly profound, one should try to get the heart rate under control quickly.
Oral agents take time to be absorbed and are not always easy to titrate. Intravenous beta-blockers such as metoprolol (Lopressor) and labetalol (Normodyne, Trandate) or intravenous diltiazem (Cardizem) can slow the heart rate quickly and can be titrated. Once the heart rate is controlled, the oral form can be started, to allow weaning from the intravenous agent. In acute management, we seek a heart rate of less than about 100 to 110 beats per minute.
If the patient’s blood pressure is marginal, loading with intravenous digoxin may be considered. The dosage is 0.5 mg intravenously, then 0.25 mg intravenously in the first 6 hours and another 0.25 mg intravenously in another 6 hours. In patients with renal insufficiency the dosage should be less, or digoxin should be avoided altogether. Often, the blood pressure will improve once the heart rate is decreased, allowing other agents to be initiated. However, if the patient is frankly hypotensive with chest pain, shortness of breath, or a diminished level of consciousness, then emergency electrical cardioversion is indicated even if anticoagulation has not yet been started (more about anticoagulation below).
Oral forms of these same agents are the workhorses for heart rate control in the outpatient setting. Oral beta-blockers and nondihydropyridine calcium channel blockers (ie, diltiazem or verapamil [Calan, Verelan]) are the first-line agents, because when digoxin is used alone, it is relatively poor at controlling the heart rate, especially when the patient is not at rest.
The choice between these agents should be dictated by whether the patient has comorbidities such as coronary artery disease, heart failure, or reactive airway disease. Nondihydropyridine calcium channel blockers are relatively contraindicated in patients with heart failure, while beta-blockers can exacerbate reactive airway disease.6
It is also important to document that the heart rate is adequately controlled outside the hospital or outpatient clinic, where the patient is typically sitting or supine. This can be done with a 6-minute walk, exercise test, or Holter monitor once rate-controlling agents have been titrated.7
When to try to restore sinus rhythm
When atrial fibrillation is first diagnosed, it may not be possible to determine if it is paroxysmal (ie, self-terminating) or persistent. If the episode does not quickly end on its own, consideration may be given to restoring sinus rhythm.
Although experts debate the merits of a rate control approach vs a rhythm control approach for managing atrial fibrillation in the long term, many, including ourselves, recommend trying to restore sinus rhythm at least once when atrial fibrillation is first discovered. It is not always clear if atrial fibrillation is truly asymptomatic. Symptoms such as fatigue or decreased exercise tolerance can be subtle. Additionally, these symptoms may be attributed to other factors such as deconditioning, obesity, or advancing age. Thus, in many cases, only restoring normal sinus rhythm for a time allows the patient and clinician to fully assess the symptoms attributable to atrial fibrillation.
Therefore, in patients with newly diagnosed atrial fibrillation, an attempt to restore sinus rhythm is often warranted. Exceptions are in select patients who have no apparent symptoms and who are very old or are deemed too frail to tolerate cardioversion.
Direct-current cardioversion is typically the treatment of choice when attempting to restore sinus rhythm. The procedure can be done without anticoagulation within 48 hours of the onset of atrial fibrillation, if the time of onset is clear.7 However, clinicians must be careful in defining the onset of atrial fibrillation for this purpose.
Symptoms such as fatigue or shortness of breath can be vague in terms of the exact time of onset and often cannot be relied upon for the purpose of deciding whether cardioversion can be done without anticoagulation. When in doubt, it is best to err on the side of safety and assume that the atrial fibrillation has been going on for more than 48 hours.
If the time of onset is unclear or if more than 48 hours have passed, there are two general strategies for proceeding to electrical cardioversion.
One is to order transesophageal echocardiography and begin anticoagulation therapy at the same time. If there is no thrombus in the left atrium, then cardioversion can be done.8 Therapeutic anticoagulation with heparin, low-molecular-weight heparin, or warfarin (Coumadin) should be achieved within 24 to 48 hours of transesophageal echocardiography and cardioversion to minimize the risk of thromboembolic events, which can occur even after sinus rhythm has been restored.
At our institution, we typically strive to achieve therapeutic anticoagulation with either heparin or low-molecular-weight heparin before cardioversion in this scenario so as to avoid situations in which a patient may undergo cardioversion but then fail to achieve therapeutic anticoagulation for some time due to unforeseen factors.
The other approach is to start warfarin and maintain a goal international normalized ratio (INR) of 2 to 3 for 3 weeks, at which time cardioversion can be performed safely without transesophageal echocardiography.8
Regardless of which strategy is used, anticoagulation should be continued for at least 4 weeks after cardioversion,8 as atrial dysfunction and the risk of stroke may persist for days to weeks after normal sinus rhythm is restored.9
Role of antiarrhythmic drugs
Antiarrhythmic drugs can be used for chemical cardioversion or, more often, to help maintain sinus rhythm after direct-current cardioversion.
Because most of these drugs have at least a small chance of restoring normal sinus rhythm, we need to follow the same rules when starting them as when performing direct-current cardioversion. Patients should not be started on an antiarrhythmic medication until they have had adequate anticoagulation for at least 3 weeks or adequate anticoagulation and a transesophageal echocardiogram confirming that there is no thrombus in the left atrium.
Antiarrhythmic drugs should be started in select patients with newly diagnosed atrial fibrillation in whom a rhythm control strategy will be pursued. For patients whose history suggests a single episode, or episodes that previously self-terminated, an antiarrhythmic may not be necessary. For those with frequent episodes or whose history suggests ongoing atrial fibrillation for a long period, an antiarrhythmic will likely be required to provide a reasonable chance of achieving freedom from atrial fibrillation.
The choice of antiarrhythmic drug should be tailored to the specific patient.
Propafenone (Rythmol) and flecainide (Tambocor) are first-line drugs7 but are contraindicated in patients with coronary artery disease and significant structural heart disease.10
Sotalol (Betapace) and dofetilide (Tikosyn) can be used in patients with coronary artery disease. However, sotalol is contraindicated in patients with congestive heart failure, and dofetilide carries a long list of drug interactions. Both must be used with extreme caution in patients with renal insufficiency, and hospital admission is required for initiation or upward titration of the dose.
Amiodarone (Cordarone) is effective, and in the short term it is typically very well tolerated. However, it has a long half-life, and its potential for long-term toxicity often makes it a poor choice for long-term treatment. The toxicity of amiodarone increases with the cumulative dose. Therefore, this drug should be avoided for long-term therapy of atrial fibrillation in younger patients.
The ‘pill-in-the-pocket’ strategy
The “pill-in-the-pocket” strategy, in which patients are instructed to take their medication only when they have a bout of atrial fibrillation, is a reasonable option for patients with symptomatic recurrences of paroxysmal atrial fibrillation. This strategy is mainly reserved for patients who have relatively infrequent recurrences. Those who have frequent recurrences are usually more effectively treated with daily dosing of an antiarrhythmic. Flecainide and propafenone are the agents of choice for this approach because of their safety profile and efficacy in chemical cardioversion.
While this strategy may be started on an outpatient basis in patients with lone, paroxysmal atrial fibrillation, those with structural heart disease or conduction abnormalities should be observed in the hospital during initiation of therapy to observe for excessive PR prolongation or development of dangerous or worrisome arrhythmias.11–13
Additionally, these agents can decrease the refractory period of the atrioventricular node, thereby increasing the ventricular rate. In the case of atrial flutter, patients may be converted from variable or 2:1 response to a 1:1 conduction. Thus, one should consider also using a beta-blocker with this strategy.
Since the goal of this approach is to convert the patient to sinus rhythm within a few hours of the onset of atrial fibrillation, it may be implemented without the use of warfarin. Patients are instructed that if they do not convert to normal sinus rhythm within a few hours, they should notify the physician so they can undergo electrical cardioversion within the 48-hour window from the onset of atrial fibrillation.
Dronedarone, a new antiarrhythmic drug
Dronedarone (Multaq) is now available and has been shown to be effective in treating atrial fibrillation.14 It has a long half-life and a mechanism of action similar to that of amiodarone. However, it may be inferior to amiodarone in terms of efficacy.15 It is metabolized by CYP3A4. No dosage adjustment is needed for patients with renal insufficiency.
Because dronedarone lacks the iodine moiety found in amiodarone, it should not carry the same toxicity profile. No pulmonary or thyroid toxicity was reported in early trials.16
Nevertheless, dronedarone has several important limitations. It carries a black box warning stating it is contraindicated in patients with severe or recently decompensated heart failure, as the mortality rate was twice as high when this drug was used in such patients in initial studies.17 Additionally, there have been reports of hepatotoxicity requiring liver transplantation in a small number of patients. The extent of this problem and strategies for avoiding it are not yet defined as of the writing of this paper. As with any new medication, patients who are started on dronedarone should be observed closely for any side effects, and these should be reported to assist in the development of the drug’s safety profile.
Pulmonary vein isolation
In a procedure that can potentially cure atrial fibrillation, catheters are inserted into the left atrium and rings of scar tissue are created around the ostia of the pulmonary veins using radiofrequency energy, electrically isolating them from the rest of the left atrium.
Some debate exists as to whether this procedure may be reasonable as a first-line therapy for some patients with atrial fibrillation.18,19 It may be considered as an early treatment strategy in a small subset of patients, specifically young patients with symptomatic, recurrent atrial fibrillation, especially if they are averse to long-term antiarrhythmic therapy.
Because patients may still be more prone to atrial arrhythmias for several weeks to months after the procedure, they must be able to tolerate anticoagulation with warfarin for at least several months.
Rate control vs rhythm control
The choice between a rate control strategy or a rhythm control strategy in the long term is not always straightforward. While atrial fibrillation is clearly associated with higher morbidity and mortality rates, there are few data to date showing that restoring and maintaining sinus rhythm in patients with atrial fibrillation reduce the incidence of morbid complications or the likelihood of death.
Thus, current guidelines recommend a rate control strategy in patients who have no symptoms, and a rhythm control strategy if rate control cannot be achieved or if symptoms persist despite adequate control of the heart rate.7 The circumstances and preferences of the individual patient should carry weight in this decision.
Trials are under way that may shed more light on the relative benefits of rhythm control with ablation or antiarrhythmics and rate control.
PREVENTING THROMBOEMBOLIC EVENTS
Warfarin
In the short term, warfarin therapy may be dictated by plans to restore sinus rhythm. Patients need warfarin for at least 4 weeks after cardioversion unless it is performed within 48 hours of the onset of atrial fibrillation.
The CHADS2score (1 point each for congestive heart failure, hypertension, age 75 or older, and diabetes mellitus; 2 points for prior stroke or transient ischemic attack) is useful when deciding whether to give long-term anticoagulation.
For patients with a score of 0, the risk of stroke is lower than the risk of a major bleeding complication while on therapeutic warfarin.20,21 For these patients, aspirin 81 to 325 mg daily is recommended for stroke prophylaxis.
For those with a score of 2 or greater, the risk of stroke without warfarin is greater than the risk of a major bleeding complication with warfarin. These patients should receive warfarin with a goal INR of 2.0 to 3.0.7
Patients with a CHADS2 score of 1 present a dilemma, as their risk of stroke without warfarin is about the same as their risk of a major bleeding complication with warfarin. They can be managed with either warfarin or aspirin, according to the physician’s judgment.7 In these cases, factors such as hobbies or professions that might increase the risk of bleeding, perceived frequency of atrial fibrillation episodes, and even patient preconceptions about warfarin are often used when deciding between aspirin and warfarin.
Patients with a CHADS2 score of 2 or greater with a single episode of atrial fibrillation and a likely reversible cause may also pose a dilemma when deciding whether to start warfarin. These patients have demonstrated they at least have the substrate to maintain atrial fibrillation. This situation again calls for physician judgment. Bear in mind that asymptomatic recurrences are common in patients with atrial fibrillation.22,23 A higher CHADS2 score denotes a greater risk of stroke and may influence this decision. It is usually beneficial to enlist the patient in this decision-making process, as patients often have very strong opinions about tolerance of the risk of stroke or of warfarin therapy itself.
Another strategy is to start anticoagulation with warfarin and aggressively monitor for recurrences. If the patient has no recurrences of atrial fibrillation after 6 to 12 months and the reversible cause is resolved, one can then revisit the need for warfarin.
Role of aspirin and clopidogrel
Aspirin, alone or in conjunction with clopidogrel (Plavix), may provide an alternative for stroke prophylaxis in patients in whom warfarin is contraindicated. While inferior to warfarin, the combination of aspirin and clopidogrel has been shown to decrease the incidence of major thromboembolic events, especially stroke.24 However, the risk of a major bleeding complication was also significantly increased.
This combination may be a reasonable strategy in select patients with a CHADS2 score of 2 or greater in whom warfarin cannot be used for reasons such as personal aversion to the medication, side effects, or nonbleeding complications or in patients whose INR is exceedingly difficult to keep within the therapeutic range.
Dabigatran, a new anticoagulant
The newest option for anticoagulation in patients with atrial fibrillation is a direct thrombin inhibitor, dabigatran (Pradaxa).
In the Randomized Evaluation of Long-Term Anticoagulation Therapy (RE-LY) trial,25 dabigatran was studied head-to-head with warfarin. The doses of dabigatran studied were 110 mg and 150 mg twice a day. At 150 mg twice a day, patients on dabigatran had a lower rate of stroke than with warfarin (1.11% vs 1.69%, P < .001), as well as a lower rate of central nervous system bleeding (0.10% vs 0.38% with warfarin, P < .001). The rates of major bleeding were comparable in the patients receiving warfarin or dabigatran 150 mg twice a day, but the rate of gastrointestinal bleeding was higher in the dabigatran group (1.51% vs 1.02% with warfarin, P < .001).25
Dabigatran was recently approved by the US Food and Drug Administration for use in patients with atrial fibrillation. Doses of 150 mg and 75 mg are available.
Dabigatran is renally excreted, and the 150 mg twice-a-day dosing is intended for patients with a creatinine clearance greater than 30 mL/min. The 75-mg twice-a-day dosing is intended for patients with a creatinine clearance of 15 to 30 mL/min. However, it should be noted that currently there are no data to support the 75-mg twice-a-day dosing.
Dabigatran does have several advantages over warfarin. Patients do not need to avoid foods containing vitamin K, and routine serial monitoring does not appear to be needed. As with any new medication, patients who are started on dabigatran should be observed closely for any side effects, and these should be reported to assist in the development of the drug’s safety profile.
SPECIAL CIRCUMSTANCES
After cardiac or noncardiac surgery
Atrial fibrillation is common after open heart surgery, occurring in approximately 25% to 50% of patients.26–28
When this happens, at least one or two attempts are made to restore sinus rhythm. Especially in the early postoperative period, anticoagulation with heparin or warfarin may be contraindicated, so careful attention must be paid to the patient’s heart rhythm so that atrial fibrillation can be recognized quickly and cardioversion performed within a 48-hour window of onset. Beta-blockers, diltiazem, and verapamil are typically used for rate control.
When atrial fibrillation recurs in patients who have undergone open heart surgery, antiarrhythmics are started early to help prevent further recurrences. At our institution, we usually use amiodarone, as it is highly effective and well tolerated in the short term. When started on amiodarone for postoperative atrial fibrillation, patients are informed that the drug will be stopped after about 2 to 3 months. For patients who continue to have bouts of atrial fibrillation, the need for antiarrhythmic medications can be reassessed, and, if needed, the optimal antiarrhythmic medication for long-term therapy for the patient can be chosen.
Atrial fibrillation in severe, acute illness
Atrial fibrillation is common in the setting of extreme systemic stressors such as shock and sepsis and when the patient is being supported with inotropic agents. In this setting, patients may be in a high-catecholamine state, and both the heart rate and the heart rhythm may be very difficult to control.
Beta-blockers and nondihydropyridine calcium channel blockers should not be used when patients are on medications to support blood pressure, and in this setting, when the patient’s hemodynamic status permits the use of these agents, their effect may be minimal.
Amiodarone or perhaps digoxin may slow the heart rate somewhat without too much effect on the blood pressure. However, with amiodarone, one may have to accept a risk of chemical cardioversion.
Electrical cardioversion with or without the assistance of an antiarrhythmic drug may control the heart rate by restoring sinus rhythm. However, atrial fibrillation often recurs, and if it recurs quickly one may have to accept elevated heart rates until the underlying process is addressed.
- Go AS, Hylek EM, Phillips KA, et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the Anticoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA 2001; 285:2370–2375.
- Lloyd-Jones DM, Wang TJ, Leip EP, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation 2004; 110:1042–1046.
- Benjamin EJ, Wolf PA, D’Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation 1998; 98:946–952.
- Kannel WB, Wolf PA, Benjamin EJ, Levy D. Prevalence, incidence, prognosis, and predisposing conditions for atrial fibrillation: population-based estimates. Am J Cardiol 1998; 82( 8A):2N–9N.
- Kannel WB, Abbott RD, Savage DD, McNamara PM. Epidemiologic features of chronic atrial fibrillation: the Framingham study. N Engl J Med 1982; 306:1018–1022.
- The Multicenter Diltiazem Postinfarction Trial Research Group. The effect of diltiazem on mortality and reinfarction after myocardial infarction. N Engl J Med 1988; 319:385–392.
- European heart Rhythm Association; Heart Rhythm society, Fuster V, Rydén LE, Cannom DS, et al. ACC/AHA/ESC guidelines for the management of patients with atrial fibrillation—executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation). J Am Coll Cardiol 2006; 48:854–906.
- Klein AL, Grimm RA, Murray RD, et al; Assessment of Cardioversion Using Transesophageal Echocardiography Investigators. Use of transesophageal echocardiography to guide cardioversion in patients with atrial fibrillation. N Engl J Med 2001; 344:1411–1120.
- Grimm RA, Leung DY, Black IW, Stewart WJ, Thomas JD, Klein AL. Left atrial appendage “stunning” after spontaneous conversion of atrial fibrillation demonstrated by transesophageal Doppler echocardiography. Am Heart J 1995; 130:174–176.
- The Cardiac Arrhythmia Suppression Trial (CAST) Investigators. Preliminary report: effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. N Engl J Med 1989; 321:406–412.
- Alboni P, Tomasi C, Menozzi C, et al. Efficacy and safety of out-of-hospital self-administered single-dose oral drug treatment in the management of infrequent, well-tolerated paroxysmal supraventricular tachycardia. J Am Coll Cardiol 2001; 37:548–553.
- Capucci A, Villani GQ, Piepoli MF. Reproducible efficacy of loading oral propafenone in restoring sinus rhythm in patients with paroxysmal atrial fibrillation. Am J Cardiol 2003; 92:1345–1347.
- Khan IA. Single oral loading dose of propafenone for pharmacological cardioversion of recent-onset atrial fibrillation. J Am Coll Cardiol 2001; 37:542–547.
- Singh BN, Connolly SJ, Crijns HJ, et al; EURIDIS and ADONIS Investigators. Dronedarone for maintenance of sinus rhythm in atrial fibrillation or flutter. N Engl J Med 2007; 357:987–999.
- Le Heuzey J, De Ferrari GM, Radzik D, Santini M, Zhu J, Davy JM. A short-term, randomized, double-blind, parallel-group study to evaluate the efficacy and safety of dronedarone versus amiodarone in patients with persistent atrial fibrillation: the DIONYSOS study. J Cardiovasc Electrophysiol 2010; 21:597–605.
- Hohnloser SH, Crijns HJ, van Eickels M, et al. Effect of dronedarone on cardiovascular events in atrial fibrillation. N Engl J Med 2009; 360:668–678.
- Køber L, Torp-Pedersen C, McMurray JJ, et al; Dronedarone Study Group. Increased mortality after dronedarone therapy for severe heart failure. N Engl J Med 2008; 358:2678–2687.
- Pappone C, Rosanio S, Augello G, et al. Mortality, morbidity, and quality of life after circumferential pulmonary vein ablation for atrial fibrillation: outcomes from a controlled nonrandomized long-term study. J Am Coll Cardiol 2003; 42:185–197.
- Wazni OM, Marrouche NF, Martin DO, et al. Radiofrequency ablation vs antiarrhythmic drugs as first-line treatment of symptomatic atrial fibrillation: a randomized trial. JAMA 2005; 293:2634–2640.
- van Walraven C, Hart RG, Singer DE, et al. Oral anticoagulants vs aspirin in nonvalvular atrial fibrillation: an individual patient metaanalysis. JAMA 2002; 288:2441–2448.
- Hart RG, Benavente O, McBride R, Pearce LA. Antithrombotic therapy to prevent stroke in patients with atrial fibrillation: a metaanalysis. Ann Intern Med 1999; 131:492–501.
- Page RL, Wilkinson WE, Clair WK, McCarthy EA, Pritchett EL. Asymptomatic arrhythmias in patients with symptomatic paroxysmal atrial fibrillation and paroxysmal supraventricular tachycardia. Circulation 1994; 89:224–227.
- Savelieva I, Camm AJ. Clinical relevance of silent atrial fibrillation: prevalence, prognosis, quality of life, and management. J Intervent Card Electrophysiol 2000; 4:369–382.
- ACTIVE Investigators, Connolly SJ, Pogue J, Hart RG, et al. Effect of clopidogrel added to aspirin in patients with atrial fibrillation. N Engl J Med 2009; 360:2066–2078.
- Connolly SJ, Ezekowitz MD, Yusuf S, et al; RE-LY Steering Committee and Investigators. Dabigatran versus warfarin in patients with atrial fibrillation. N Engl J Med 2009; 361:1139–1151. Erratum in: N Engl J Med 2010; 363:1877.
- Almassi GH, Schowalter T, Nicolosi AC, et al. Atrial fibrillation after cardiac surgery: a major morbid event? Ann Surg 1997; 226:501–511.
- Creswell LL, Schuessler RB, Rosenbloom M, Cox JL. Hazards of postoperative atrial arrhythmias. Ann Thorac Surg 1993; 56:539–549.
- Mathew JP, Fontes ML, Tudor IC, et al; Investigators of the Ischemia Research and Education Foundation; Multicenter Study of Perioperative Ischemia Research Group. A multicenter risk index for atrial fibrillation after cardiac surgery. JAMA 2004; 291:1720–1729.
- Go AS, Hylek EM, Phillips KA, et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the Anticoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA 2001; 285:2370–2375.
- Lloyd-Jones DM, Wang TJ, Leip EP, et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation 2004; 110:1042–1046.
- Benjamin EJ, Wolf PA, D’Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation 1998; 98:946–952.
- Kannel WB, Wolf PA, Benjamin EJ, Levy D. Prevalence, incidence, prognosis, and predisposing conditions for atrial fibrillation: population-based estimates. Am J Cardiol 1998; 82( 8A):2N–9N.
- Kannel WB, Abbott RD, Savage DD, McNamara PM. Epidemiologic features of chronic atrial fibrillation: the Framingham study. N Engl J Med 1982; 306:1018–1022.
- The Multicenter Diltiazem Postinfarction Trial Research Group. The effect of diltiazem on mortality and reinfarction after myocardial infarction. N Engl J Med 1988; 319:385–392.
- European heart Rhythm Association; Heart Rhythm society, Fuster V, Rydén LE, Cannom DS, et al. ACC/AHA/ESC guidelines for the management of patients with atrial fibrillation—executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation). J Am Coll Cardiol 2006; 48:854–906.
- Klein AL, Grimm RA, Murray RD, et al; Assessment of Cardioversion Using Transesophageal Echocardiography Investigators. Use of transesophageal echocardiography to guide cardioversion in patients with atrial fibrillation. N Engl J Med 2001; 344:1411–1120.
- Grimm RA, Leung DY, Black IW, Stewart WJ, Thomas JD, Klein AL. Left atrial appendage “stunning” after spontaneous conversion of atrial fibrillation demonstrated by transesophageal Doppler echocardiography. Am Heart J 1995; 130:174–176.
- The Cardiac Arrhythmia Suppression Trial (CAST) Investigators. Preliminary report: effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. N Engl J Med 1989; 321:406–412.
- Alboni P, Tomasi C, Menozzi C, et al. Efficacy and safety of out-of-hospital self-administered single-dose oral drug treatment in the management of infrequent, well-tolerated paroxysmal supraventricular tachycardia. J Am Coll Cardiol 2001; 37:548–553.
- Capucci A, Villani GQ, Piepoli MF. Reproducible efficacy of loading oral propafenone in restoring sinus rhythm in patients with paroxysmal atrial fibrillation. Am J Cardiol 2003; 92:1345–1347.
- Khan IA. Single oral loading dose of propafenone for pharmacological cardioversion of recent-onset atrial fibrillation. J Am Coll Cardiol 2001; 37:542–547.
- Singh BN, Connolly SJ, Crijns HJ, et al; EURIDIS and ADONIS Investigators. Dronedarone for maintenance of sinus rhythm in atrial fibrillation or flutter. N Engl J Med 2007; 357:987–999.
- Le Heuzey J, De Ferrari GM, Radzik D, Santini M, Zhu J, Davy JM. A short-term, randomized, double-blind, parallel-group study to evaluate the efficacy and safety of dronedarone versus amiodarone in patients with persistent atrial fibrillation: the DIONYSOS study. J Cardiovasc Electrophysiol 2010; 21:597–605.
- Hohnloser SH, Crijns HJ, van Eickels M, et al. Effect of dronedarone on cardiovascular events in atrial fibrillation. N Engl J Med 2009; 360:668–678.
- Køber L, Torp-Pedersen C, McMurray JJ, et al; Dronedarone Study Group. Increased mortality after dronedarone therapy for severe heart failure. N Engl J Med 2008; 358:2678–2687.
- Pappone C, Rosanio S, Augello G, et al. Mortality, morbidity, and quality of life after circumferential pulmonary vein ablation for atrial fibrillation: outcomes from a controlled nonrandomized long-term study. J Am Coll Cardiol 2003; 42:185–197.
- Wazni OM, Marrouche NF, Martin DO, et al. Radiofrequency ablation vs antiarrhythmic drugs as first-line treatment of symptomatic atrial fibrillation: a randomized trial. JAMA 2005; 293:2634–2640.
- van Walraven C, Hart RG, Singer DE, et al. Oral anticoagulants vs aspirin in nonvalvular atrial fibrillation: an individual patient metaanalysis. JAMA 2002; 288:2441–2448.
- Hart RG, Benavente O, McBride R, Pearce LA. Antithrombotic therapy to prevent stroke in patients with atrial fibrillation: a metaanalysis. Ann Intern Med 1999; 131:492–501.
- Page RL, Wilkinson WE, Clair WK, McCarthy EA, Pritchett EL. Asymptomatic arrhythmias in patients with symptomatic paroxysmal atrial fibrillation and paroxysmal supraventricular tachycardia. Circulation 1994; 89:224–227.
- Savelieva I, Camm AJ. Clinical relevance of silent atrial fibrillation: prevalence, prognosis, quality of life, and management. J Intervent Card Electrophysiol 2000; 4:369–382.
- ACTIVE Investigators, Connolly SJ, Pogue J, Hart RG, et al. Effect of clopidogrel added to aspirin in patients with atrial fibrillation. N Engl J Med 2009; 360:2066–2078.
- Connolly SJ, Ezekowitz MD, Yusuf S, et al; RE-LY Steering Committee and Investigators. Dabigatran versus warfarin in patients with atrial fibrillation. N Engl J Med 2009; 361:1139–1151. Erratum in: N Engl J Med 2010; 363:1877.
- Almassi GH, Schowalter T, Nicolosi AC, et al. Atrial fibrillation after cardiac surgery: a major morbid event? Ann Surg 1997; 226:501–511.
- Creswell LL, Schuessler RB, Rosenbloom M, Cox JL. Hazards of postoperative atrial arrhythmias. Ann Thorac Surg 1993; 56:539–549.
- Mathew JP, Fontes ML, Tudor IC, et al; Investigators of the Ischemia Research and Education Foundation; Multicenter Study of Perioperative Ischemia Research Group. A multicenter risk index for atrial fibrillation after cardiac surgery. JAMA 2004; 291:1720–1729.
KEY POINTS
- When atrial fibrillation is newly diagnosed, reversible causes and commonly associated processes should be sought.
- Agents to control the heart rate, eg, beta-blockers or nondihydropyridine calcium channel blockers, are often started and titrated intravenously and then changed to oral dosing.
- The benefit of rhythm control has not been firmly established. Although we try cardioversion at least once when atrial fibrillation is first diagnosed, rhythm control is generally reserved for patients whose symptoms persist despite rate control, or for patients in whom the heart rate cannot be controlled.
- The need for short-term or long-term anticoagulation must be estimated.
Perioperative Medicine Summit 2011
Summit Director:
Amir K. Jaffer, MD
Contents
Abstract 1: Application of 2007 ACC/AHA guidelines on perioperative cardiovascular evaluation and care for noncardiac surgery using decision support tools
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 2: Prevalence of obstructive dleep spnea in patients presenting for hip or knee replacement surgery
Micah Beachy, DO; Jason Shiffermiller, MD; and Chad Vokoun, MD
Abstract 3: A protocol to triage preoperative assessments to either nurses or nurse practitioners/physician assistants
Anthony Basil, RN; Pamela Pennigar, FNP; David R. Wright, MD; and Ronald P. Olson, MD
Abstract 4: Application of 2007 ACC/AHA guidelines on perioperative cardiovascular evaluation and care for noncardiac surgery using decision support tools
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 5: Most anesthesiologists don’t correctly apply 2007 ACC/AHA guidelines on perioperative cardiac evaluation
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, Kris Arheart, and Richard Dutton
Abstract 6: Anesthesiology residents do not agree with their training programs on the degree to which the 2007 ACC/AHA guidelines are emphasized
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 7: Prevalence of obstructive sleep apnea in patients presenting for hip or knee replacement surgery
Micah Beachy, DO; Jason Shiffermiller, MD; and Chad Vokoun, MD
Abstract 8: A protocol to triage preoperative assessments to either nurses or nurse practitioners/physician assistants
Anthony Basil, RN; Pamela Pennigar, FNP; David R. Wright, MD; and Ronald P. Olson, MD
Abstract 9: Do ACEIs on the morning of surgery increase risk of intraoperative hypotension?
Steven L. Cohn, MD, and Kalia Skeete, MD
Abstract 10: One-year incidence of postoperative troponin revations in patients undergoing major orthopedic surgery
Michael Urban, MD, PhD; Stephen Wolfe, BS; Niel Sanghevi, BS; and Steven Magid, MD
Abstract 11: A review of preoperative clinic cardiology referrals for adults undergoing intermediate- and low-risk surgery
Susan Calderwood, MD; Jennifer Lee Morse, MS; and Damon R. Michaels, CCRP
Abstract 12: Patterns of preoperative consultation by risk and surgical specialty in a large health care system
Stephan Thilen, MD, MS; Christopher Bryson, MD, MS; Robert Reid, MD, PhD; and Miriam Treggiari, MD, MPH, PhD
Abstract 13: One-year incidence for admission to a critical care unit after major orthopedic surgery
Michael Urban, MD, PhD; Steven Magid, MD; and Michele Mangini, DNP
Abstract 14: Determination of the causes of long patient wait times in a preoperative evaluation clinic
Jean Kwo, MD; Devon Price, BS; Mary Elizabeth Ellbeg, RN; and Retsef Levi, PhD
Abstract 15: Does perioperative statin treatment affect hospital and ICU length of stay rollowing cardiac surgery: A systematic review
Vineet Chopra, MD, FACP, FHM; David Wesorick, MD; and Kim A. Eagle, MD
Abstract 16: Assessment of patient satisfaction of nurse screening vs complete preoperative assessment
Ronald Olson, MD, and Kathy Bock, RN
Abstract 17: Traumatic subdural hematoma: An update on morbidity
Rachel Thompson, MD; Christina Ryan, MD; Nancy Temkin, PhD; Richard Ellenbogen, MD; and Joann G. Elmore, MD, MPH
Abstract 18: Lipid emulsion as a lifesaving treatment for local anesthetic systemic toxicity (LAST)
Deepti Sachdev and Guy Weinberg, MD
Abstract 19: Perioperative ACLS recommendations should be modified for the treatment of local anesthetic toxicity
Adam Haas, MD, and Alexia Beccue, MD
Abstract 20: Preoperative EMR containing smart-set reminders improve accuracy of documentation by nonanesthesia clinicians during preoperative assessments
Angela Edwards, MD; Jill Grant, PA; and Ruth Hyde, MD
Abstract 21: POET: Procedure outcomes evaluation tool
Ahmad AbuSalah, MSc, and Terrence Adam, MD, PhD
Abstract 22: Results of a multidisciplinary preoperative assessment process for high-risk orthopedic patients
Terrence Adam, MD, PhD; Connie Parenti, MD; Terence Gioe, MD; and Karen Ringsred, MD
Abstract 23: Practical algorithm for preoperative evaluation of patients with liver disease
Madalina A. Vlase, PA-C, and Deborah C. Richman, MBChB, FFA(SA)
Abstract 24: Evaluation and management of isolated elevated aPTT
Sheila Hassan, MSN, NP; Patricia Kidik, MSN, NP; Catherine McGowan, MSN, NP; and Angela M. Bader, MD
Abstract 25: A perioperative triage plan for obstructive sleep apnea patients
Christian Altman, MD; R. Michael Boyer, DO; and Peter G. Kallas, MD
Abstract 26: Quantitative evaluation of handoff checklists
Jay Joshi, MD, and David Mayer, MD
Abstract 27: To deflate or not to deflate: Lap-Band® management in subsequent surgeries
Arjun Reddy, MD, and Deborah C. Richman, MBChB, FFA(SA)
Abstract 28: Takotsubo cardiomyopathy and resultant cardiogenic shock after mitral valve repair
Adam Evans, MD, MBA; Daniel B. Sims, MD; Nir Uriel, MD; Ulrich P. Jorde, MD; and Craig R. Smith, MD
Abstract 29: Intravenous vitamin K: Rapid reversal of warfarin and lack of subsequent warfarin resistance
Feras Abdul Khalek, MD; Interdeep Dhaliwal, MD; and Twylla Tassava, MD
Abstract 30: Cervical spine surgery: When not to extubate postoperatively
Carlos Mateo Mijares, MD; Doris Debs, ARNP, MSN-BC; Nicole Martin, MD; and Ronald Lee Samson, MD
Abstract 31: Total occlusion of oral cavity by mandibular sarcoma for resection: To intubate nasally or proceed to an awake tracheostomy?
Carlos Mateo Mijares, MD, and Maria DeLapena, MD
Abstract 32: Perioperative fatal embolic stroke associated with iron deficiency anemia and thrombocytosis
Carlos Mateo Mijares, MD; Nicole Martin, MD; and Ricardo Martinez-Ruiz, MD
Abstract 33: Conservative approach saves the day anesthesia-wise and surgical-wise
Carlos Mateo Mijares, MD; Bradley Shore, MD; Edward Zalkind, CRNA; and Nicole Martin, MD
Abstract 34: Predictors of acute kidney injury in patients undergoing total knee replacement surgery
Vishal Sehgal, MD; Pardeep Bansal, MD; Praveen Reddy, MD; Vishal Sharma, MD; Samuel Lesko, MD; John H. Doherty, MD; Theodore Tomaszewski, MD; Jack Prior, MD; Roger Getts, MD; and Jeremiah Eagan, MD
Abstract 35: Perioperative medical management of the Marfan patient undergoing repeat cardiothoracic surgery
Aashish Shah, MD, and Adam Skrzynski, MD
Summit Director:
Amir K. Jaffer, MD
Contents
Abstract 1: Application of 2007 ACC/AHA guidelines on perioperative cardiovascular evaluation and care for noncardiac surgery using decision support tools
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 2: Prevalence of obstructive dleep spnea in patients presenting for hip or knee replacement surgery
Micah Beachy, DO; Jason Shiffermiller, MD; and Chad Vokoun, MD
Abstract 3: A protocol to triage preoperative assessments to either nurses or nurse practitioners/physician assistants
Anthony Basil, RN; Pamela Pennigar, FNP; David R. Wright, MD; and Ronald P. Olson, MD
Abstract 4: Application of 2007 ACC/AHA guidelines on perioperative cardiovascular evaluation and care for noncardiac surgery using decision support tools
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 5: Most anesthesiologists don’t correctly apply 2007 ACC/AHA guidelines on perioperative cardiac evaluation
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, Kris Arheart, and Richard Dutton
Abstract 6: Anesthesiology residents do not agree with their training programs on the degree to which the 2007 ACC/AHA guidelines are emphasized
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 7: Prevalence of obstructive sleep apnea in patients presenting for hip or knee replacement surgery
Micah Beachy, DO; Jason Shiffermiller, MD; and Chad Vokoun, MD
Abstract 8: A protocol to triage preoperative assessments to either nurses or nurse practitioners/physician assistants
Anthony Basil, RN; Pamela Pennigar, FNP; David R. Wright, MD; and Ronald P. Olson, MD
Abstract 9: Do ACEIs on the morning of surgery increase risk of intraoperative hypotension?
Steven L. Cohn, MD, and Kalia Skeete, MD
Abstract 10: One-year incidence of postoperative troponin revations in patients undergoing major orthopedic surgery
Michael Urban, MD, PhD; Stephen Wolfe, BS; Niel Sanghevi, BS; and Steven Magid, MD
Abstract 11: A review of preoperative clinic cardiology referrals for adults undergoing intermediate- and low-risk surgery
Susan Calderwood, MD; Jennifer Lee Morse, MS; and Damon R. Michaels, CCRP
Abstract 12: Patterns of preoperative consultation by risk and surgical specialty in a large health care system
Stephan Thilen, MD, MS; Christopher Bryson, MD, MS; Robert Reid, MD, PhD; and Miriam Treggiari, MD, MPH, PhD
Abstract 13: One-year incidence for admission to a critical care unit after major orthopedic surgery
Michael Urban, MD, PhD; Steven Magid, MD; and Michele Mangini, DNP
Abstract 14: Determination of the causes of long patient wait times in a preoperative evaluation clinic
Jean Kwo, MD; Devon Price, BS; Mary Elizabeth Ellbeg, RN; and Retsef Levi, PhD
Abstract 15: Does perioperative statin treatment affect hospital and ICU length of stay rollowing cardiac surgery: A systematic review
Vineet Chopra, MD, FACP, FHM; David Wesorick, MD; and Kim A. Eagle, MD
Abstract 16: Assessment of patient satisfaction of nurse screening vs complete preoperative assessment
Ronald Olson, MD, and Kathy Bock, RN
Abstract 17: Traumatic subdural hematoma: An update on morbidity
Rachel Thompson, MD; Christina Ryan, MD; Nancy Temkin, PhD; Richard Ellenbogen, MD; and Joann G. Elmore, MD, MPH
Abstract 18: Lipid emulsion as a lifesaving treatment for local anesthetic systemic toxicity (LAST)
Deepti Sachdev and Guy Weinberg, MD
Abstract 19: Perioperative ACLS recommendations should be modified for the treatment of local anesthetic toxicity
Adam Haas, MD, and Alexia Beccue, MD
Abstract 20: Preoperative EMR containing smart-set reminders improve accuracy of documentation by nonanesthesia clinicians during preoperative assessments
Angela Edwards, MD; Jill Grant, PA; and Ruth Hyde, MD
Abstract 21: POET: Procedure outcomes evaluation tool
Ahmad AbuSalah, MSc, and Terrence Adam, MD, PhD
Abstract 22: Results of a multidisciplinary preoperative assessment process for high-risk orthopedic patients
Terrence Adam, MD, PhD; Connie Parenti, MD; Terence Gioe, MD; and Karen Ringsred, MD
Abstract 23: Practical algorithm for preoperative evaluation of patients with liver disease
Madalina A. Vlase, PA-C, and Deborah C. Richman, MBChB, FFA(SA)
Abstract 24: Evaluation and management of isolated elevated aPTT
Sheila Hassan, MSN, NP; Patricia Kidik, MSN, NP; Catherine McGowan, MSN, NP; and Angela M. Bader, MD
Abstract 25: A perioperative triage plan for obstructive sleep apnea patients
Christian Altman, MD; R. Michael Boyer, DO; and Peter G. Kallas, MD
Abstract 26: Quantitative evaluation of handoff checklists
Jay Joshi, MD, and David Mayer, MD
Abstract 27: To deflate or not to deflate: Lap-Band® management in subsequent surgeries
Arjun Reddy, MD, and Deborah C. Richman, MBChB, FFA(SA)
Abstract 28: Takotsubo cardiomyopathy and resultant cardiogenic shock after mitral valve repair
Adam Evans, MD, MBA; Daniel B. Sims, MD; Nir Uriel, MD; Ulrich P. Jorde, MD; and Craig R. Smith, MD
Abstract 29: Intravenous vitamin K: Rapid reversal of warfarin and lack of subsequent warfarin resistance
Feras Abdul Khalek, MD; Interdeep Dhaliwal, MD; and Twylla Tassava, MD
Abstract 30: Cervical spine surgery: When not to extubate postoperatively
Carlos Mateo Mijares, MD; Doris Debs, ARNP, MSN-BC; Nicole Martin, MD; and Ronald Lee Samson, MD
Abstract 31: Total occlusion of oral cavity by mandibular sarcoma for resection: To intubate nasally or proceed to an awake tracheostomy?
Carlos Mateo Mijares, MD, and Maria DeLapena, MD
Abstract 32: Perioperative fatal embolic stroke associated with iron deficiency anemia and thrombocytosis
Carlos Mateo Mijares, MD; Nicole Martin, MD; and Ricardo Martinez-Ruiz, MD
Abstract 33: Conservative approach saves the day anesthesia-wise and surgical-wise
Carlos Mateo Mijares, MD; Bradley Shore, MD; Edward Zalkind, CRNA; and Nicole Martin, MD
Abstract 34: Predictors of acute kidney injury in patients undergoing total knee replacement surgery
Vishal Sehgal, MD; Pardeep Bansal, MD; Praveen Reddy, MD; Vishal Sharma, MD; Samuel Lesko, MD; John H. Doherty, MD; Theodore Tomaszewski, MD; Jack Prior, MD; Roger Getts, MD; and Jeremiah Eagan, MD
Abstract 35: Perioperative medical management of the Marfan patient undergoing repeat cardiothoracic surgery
Aashish Shah, MD, and Adam Skrzynski, MD
Summit Director:
Amir K. Jaffer, MD
Contents
Abstract 1: Application of 2007 ACC/AHA guidelines on perioperative cardiovascular evaluation and care for noncardiac surgery using decision support tools
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 2: Prevalence of obstructive dleep spnea in patients presenting for hip or knee replacement surgery
Micah Beachy, DO; Jason Shiffermiller, MD; and Chad Vokoun, MD
Abstract 3: A protocol to triage preoperative assessments to either nurses or nurse practitioners/physician assistants
Anthony Basil, RN; Pamela Pennigar, FNP; David R. Wright, MD; and Ronald P. Olson, MD
Abstract 4: Application of 2007 ACC/AHA guidelines on perioperative cardiovascular evaluation and care for noncardiac surgery using decision support tools
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 5: Most anesthesiologists don’t correctly apply 2007 ACC/AHA guidelines on perioperative cardiac evaluation
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, Kris Arheart, and Richard Dutton
Abstract 6: Anesthesiology residents do not agree with their training programs on the degree to which the 2007 ACC/AHA guidelines are emphasized
BobbieJean Sweitzer, Michael Vigoda, Vicente Behrens, Nikola Miljkovic, and Kris Arheart
Abstract 7: Prevalence of obstructive sleep apnea in patients presenting for hip or knee replacement surgery
Micah Beachy, DO; Jason Shiffermiller, MD; and Chad Vokoun, MD
Abstract 8: A protocol to triage preoperative assessments to either nurses or nurse practitioners/physician assistants
Anthony Basil, RN; Pamela Pennigar, FNP; David R. Wright, MD; and Ronald P. Olson, MD
Abstract 9: Do ACEIs on the morning of surgery increase risk of intraoperative hypotension?
Steven L. Cohn, MD, and Kalia Skeete, MD
Abstract 10: One-year incidence of postoperative troponin revations in patients undergoing major orthopedic surgery
Michael Urban, MD, PhD; Stephen Wolfe, BS; Niel Sanghevi, BS; and Steven Magid, MD
Abstract 11: A review of preoperative clinic cardiology referrals for adults undergoing intermediate- and low-risk surgery
Susan Calderwood, MD; Jennifer Lee Morse, MS; and Damon R. Michaels, CCRP
Abstract 12: Patterns of preoperative consultation by risk and surgical specialty in a large health care system
Stephan Thilen, MD, MS; Christopher Bryson, MD, MS; Robert Reid, MD, PhD; and Miriam Treggiari, MD, MPH, PhD
Abstract 13: One-year incidence for admission to a critical care unit after major orthopedic surgery
Michael Urban, MD, PhD; Steven Magid, MD; and Michele Mangini, DNP
Abstract 14: Determination of the causes of long patient wait times in a preoperative evaluation clinic
Jean Kwo, MD; Devon Price, BS; Mary Elizabeth Ellbeg, RN; and Retsef Levi, PhD
Abstract 15: Does perioperative statin treatment affect hospital and ICU length of stay rollowing cardiac surgery: A systematic review
Vineet Chopra, MD, FACP, FHM; David Wesorick, MD; and Kim A. Eagle, MD
Abstract 16: Assessment of patient satisfaction of nurse screening vs complete preoperative assessment
Ronald Olson, MD, and Kathy Bock, RN
Abstract 17: Traumatic subdural hematoma: An update on morbidity
Rachel Thompson, MD; Christina Ryan, MD; Nancy Temkin, PhD; Richard Ellenbogen, MD; and Joann G. Elmore, MD, MPH
Abstract 18: Lipid emulsion as a lifesaving treatment for local anesthetic systemic toxicity (LAST)
Deepti Sachdev and Guy Weinberg, MD
Abstract 19: Perioperative ACLS recommendations should be modified for the treatment of local anesthetic toxicity
Adam Haas, MD, and Alexia Beccue, MD
Abstract 20: Preoperative EMR containing smart-set reminders improve accuracy of documentation by nonanesthesia clinicians during preoperative assessments
Angela Edwards, MD; Jill Grant, PA; and Ruth Hyde, MD
Abstract 21: POET: Procedure outcomes evaluation tool
Ahmad AbuSalah, MSc, and Terrence Adam, MD, PhD
Abstract 22: Results of a multidisciplinary preoperative assessment process for high-risk orthopedic patients
Terrence Adam, MD, PhD; Connie Parenti, MD; Terence Gioe, MD; and Karen Ringsred, MD
Abstract 23: Practical algorithm for preoperative evaluation of patients with liver disease
Madalina A. Vlase, PA-C, and Deborah C. Richman, MBChB, FFA(SA)
Abstract 24: Evaluation and management of isolated elevated aPTT
Sheila Hassan, MSN, NP; Patricia Kidik, MSN, NP; Catherine McGowan, MSN, NP; and Angela M. Bader, MD
Abstract 25: A perioperative triage plan for obstructive sleep apnea patients
Christian Altman, MD; R. Michael Boyer, DO; and Peter G. Kallas, MD
Abstract 26: Quantitative evaluation of handoff checklists
Jay Joshi, MD, and David Mayer, MD
Abstract 27: To deflate or not to deflate: Lap-Band® management in subsequent surgeries
Arjun Reddy, MD, and Deborah C. Richman, MBChB, FFA(SA)
Abstract 28: Takotsubo cardiomyopathy and resultant cardiogenic shock after mitral valve repair
Adam Evans, MD, MBA; Daniel B. Sims, MD; Nir Uriel, MD; Ulrich P. Jorde, MD; and Craig R. Smith, MD
Abstract 29: Intravenous vitamin K: Rapid reversal of warfarin and lack of subsequent warfarin resistance
Feras Abdul Khalek, MD; Interdeep Dhaliwal, MD; and Twylla Tassava, MD
Abstract 30: Cervical spine surgery: When not to extubate postoperatively
Carlos Mateo Mijares, MD; Doris Debs, ARNP, MSN-BC; Nicole Martin, MD; and Ronald Lee Samson, MD
Abstract 31: Total occlusion of oral cavity by mandibular sarcoma for resection: To intubate nasally or proceed to an awake tracheostomy?
Carlos Mateo Mijares, MD, and Maria DeLapena, MD
Abstract 32: Perioperative fatal embolic stroke associated with iron deficiency anemia and thrombocytosis
Carlos Mateo Mijares, MD; Nicole Martin, MD; and Ricardo Martinez-Ruiz, MD
Abstract 33: Conservative approach saves the day anesthesia-wise and surgical-wise
Carlos Mateo Mijares, MD; Bradley Shore, MD; Edward Zalkind, CRNA; and Nicole Martin, MD
Abstract 34: Predictors of acute kidney injury in patients undergoing total knee replacement surgery
Vishal Sehgal, MD; Pardeep Bansal, MD; Praveen Reddy, MD; Vishal Sharma, MD; Samuel Lesko, MD; John H. Doherty, MD; Theodore Tomaszewski, MD; Jack Prior, MD; Roger Getts, MD; and Jeremiah Eagan, MD
Abstract 35: Perioperative medical management of the Marfan patient undergoing repeat cardiothoracic surgery
Aashish Shah, MD, and Adam Skrzynski, MD