User login
New mAb can overcome resistance to other mAbs

Photo courtesy of the
University of Southampton
A newly developed monoclonal antibody (mAb) can reverse resistance to other mAbs in chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL), according to research published in Cancer Cell.
Investigators found that some cancer cells draw mAbs inside themselves, making them invisible to immune cells.
But a mAb called BI-1206 can prevent this process and enhance cancer killing by binding to a molecule called FcγRIIB.
In preclinical experiments, BI-1206 was able to overcome resistance to mAbs such as rituximab.
“With more monoclonal antibody treatments being developed, there is an urgent need to understand how tumors become resistant to them and develop ways to overcome it,” said study author Mark Cragg, PhD, of the University of Southampton in the UK.
“Not only does BI-1206 appear to be able to reverse resistance to a range of monoclonal antibodies, it is also effective at directly killing cancer cells itself.”
In the Cancer Cell paper, BI-1206 is referred to as 6G11. The investigators found that 6G11 can block rituximab internalization and has “potent antitumor activity” in vitro. 6G11 was also well-tolerated and did not prompt cytokine storm.
In a mouse model of CLL, 6G11 enhanced rituximab-mediated depletion of primary CLL cells and improved responses when compared to rituximab alone.
In mice engrafted with cells from patients with CLL that was refractory to rituximab, ofatumumab, and/or alemtuzumab, 6G11 alone depleted CLL cells but did not improve overall response rates compared to rituximab alone. However, 6G11 in combination with rituximab did improve overall response rates compared to rituximab alone.
In a mouse model of MCL, neither 6G11 nor rituximab alone improved long-term survival. However, 30% of mice treated with both drugs survived tumor-free out to 100 days.
Combining 6G11 with obinutuzumab significantly improved splenic tumor cell depletion in mice with CLL. And more than 90% of mice that received 6G11 and alemtuzumab had a complete response to the treatment.
The investigators said these data suggest 6G11 can overcome mAb resistance for multiple targets. They said the drug will be tested in patients with CLL and non-Hodgkin lymphoma in an early stage clinical trial. ![]()

Photo courtesy of the
University of Southampton
A newly developed monoclonal antibody (mAb) can reverse resistance to other mAbs in chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL), according to research published in Cancer Cell.
Investigators found that some cancer cells draw mAbs inside themselves, making them invisible to immune cells.
But a mAb called BI-1206 can prevent this process and enhance cancer killing by binding to a molecule called FcγRIIB.
In preclinical experiments, BI-1206 was able to overcome resistance to mAbs such as rituximab.
“With more monoclonal antibody treatments being developed, there is an urgent need to understand how tumors become resistant to them and develop ways to overcome it,” said study author Mark Cragg, PhD, of the University of Southampton in the UK.
“Not only does BI-1206 appear to be able to reverse resistance to a range of monoclonal antibodies, it is also effective at directly killing cancer cells itself.”
In the Cancer Cell paper, BI-1206 is referred to as 6G11. The investigators found that 6G11 can block rituximab internalization and has “potent antitumor activity” in vitro. 6G11 was also well-tolerated and did not prompt cytokine storm.
In a mouse model of CLL, 6G11 enhanced rituximab-mediated depletion of primary CLL cells and improved responses when compared to rituximab alone.
In mice engrafted with cells from patients with CLL that was refractory to rituximab, ofatumumab, and/or alemtuzumab, 6G11 alone depleted CLL cells but did not improve overall response rates compared to rituximab alone. However, 6G11 in combination with rituximab did improve overall response rates compared to rituximab alone.
In a mouse model of MCL, neither 6G11 nor rituximab alone improved long-term survival. However, 30% of mice treated with both drugs survived tumor-free out to 100 days.
Combining 6G11 with obinutuzumab significantly improved splenic tumor cell depletion in mice with CLL. And more than 90% of mice that received 6G11 and alemtuzumab had a complete response to the treatment.
The investigators said these data suggest 6G11 can overcome mAb resistance for multiple targets. They said the drug will be tested in patients with CLL and non-Hodgkin lymphoma in an early stage clinical trial. ![]()

Photo courtesy of the
University of Southampton
A newly developed monoclonal antibody (mAb) can reverse resistance to other mAbs in chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL), according to research published in Cancer Cell.
Investigators found that some cancer cells draw mAbs inside themselves, making them invisible to immune cells.
But a mAb called BI-1206 can prevent this process and enhance cancer killing by binding to a molecule called FcγRIIB.
In preclinical experiments, BI-1206 was able to overcome resistance to mAbs such as rituximab.
“With more monoclonal antibody treatments being developed, there is an urgent need to understand how tumors become resistant to them and develop ways to overcome it,” said study author Mark Cragg, PhD, of the University of Southampton in the UK.
“Not only does BI-1206 appear to be able to reverse resistance to a range of monoclonal antibodies, it is also effective at directly killing cancer cells itself.”
In the Cancer Cell paper, BI-1206 is referred to as 6G11. The investigators found that 6G11 can block rituximab internalization and has “potent antitumor activity” in vitro. 6G11 was also well-tolerated and did not prompt cytokine storm.
In a mouse model of CLL, 6G11 enhanced rituximab-mediated depletion of primary CLL cells and improved responses when compared to rituximab alone.
In mice engrafted with cells from patients with CLL that was refractory to rituximab, ofatumumab, and/or alemtuzumab, 6G11 alone depleted CLL cells but did not improve overall response rates compared to rituximab alone. However, 6G11 in combination with rituximab did improve overall response rates compared to rituximab alone.
In a mouse model of MCL, neither 6G11 nor rituximab alone improved long-term survival. However, 30% of mice treated with both drugs survived tumor-free out to 100 days.
Combining 6G11 with obinutuzumab significantly improved splenic tumor cell depletion in mice with CLL. And more than 90% of mice that received 6G11 and alemtuzumab had a complete response to the treatment.
The investigators said these data suggest 6G11 can overcome mAb resistance for multiple targets. They said the drug will be tested in patients with CLL and non-Hodgkin lymphoma in an early stage clinical trial. ![]()
Tumor sequencing fails to paint the whole picture

Image courtesy of NIGMS
Many of the genetic alterations revealed by sequencing a cancer patient’s tumor DNA are not actually associated with the cancer, according to a study published in Science Translational Medicine.
In fact, these alterations are inherited germline mutations already present in an individual’s normal cells.
To make this discovery, researchers compared DNA from tumors and normal cells in 815 patients with hematologic and solid tumor malignancies.
Almost half of the patients analyzed using tumor-only approaches had genetic alterations in their tumors that were also present in their normal cells, which suggests the alterations were “false-positive” changes not specific to the tumor.
As personalized medicine is predicated on tailoring treatments to the genetic makeup of a patient’s tumor, the high rate of false-positives uncovered in this study has implications for the accuracy of the approach when it relies on tumor-only sequencing, the researchers said.
“We knew from our pioneering whole-exome analyses of cancer patients that a significant number of the genetic alterations that were thought to be associated with tumors were also present in the inherited germline DNA,” said Siân Jones, PhD, of Personal Genome Diagnostics in Baltimore, Maryland.
“By comparing tumor DNA to DNA from normal tissue, we were able to separate out those genetic alterations that are truly tumor-specific. Accurately identifying tumor-specific alterations is essential to realizing the potential of personalized medicine to achieve better treatment outcomes.”
The researchers identified 382 genetic alterations that were potentially tumor-specific by first detecting all of the genetic changes in patients’ tumors and then eliminating those that were well-known germline alterations.
However, when the remaining alterations were compared to the genomic profiles of the patients’ germline DNA, an average of 249, or 65%, turned out to be false-positive changes that were already present in the normal cells.
The researchers also looked at the alterations in actionable genes, which have been identified as potential targets for approved or investigational cancer therapies. They found that almost half of the tumor samples (48%) had at least one false-positive mutation in an actionable gene.
The use of false-positive findings to guide personalized treatment decisions could result in a substantial number of patients receiving therapies that are not optimized for their cancer, according to the researchers. Therefore, it seems sequencing normal DNA alongside tumor DNA is essential.
In addition to selecting personalized therapies for cancer patients, sequencing normal DNA can increase the overall understanding of cancer, including finding cancer predisposition due to germline genome changes, said Victor Velculescu, MD, PhD, of Johns Hopkins University School of Medicine in Baltimore, Maryland.
In this study, the germline analyses revealed changes in cancer-related genes in 3% of the patients who had no known signs of genetically linked cancer.
“These analyses can help us find alterations in cancer-predisposing genes in ways that weren’t previously appreciated,” Dr Velculescu said.
He also acknowledged, however, that there are challenges to implementing tumor-normal DNA analyses in a clinical setting. These include the additional work and costs of sequencing and analyzing a patient’s normal tissue along with his or her tumor tissue.
Costs for tumor gene sequencing begin at several thousand dollars, which would increase if sequencing was done on DNA from normal tissue as well. And health insurance may not fully cover normal-tissue genetic sequencing. ![]()

Image courtesy of NIGMS
Many of the genetic alterations revealed by sequencing a cancer patient’s tumor DNA are not actually associated with the cancer, according to a study published in Science Translational Medicine.
In fact, these alterations are inherited germline mutations already present in an individual’s normal cells.
To make this discovery, researchers compared DNA from tumors and normal cells in 815 patients with hematologic and solid tumor malignancies.
Almost half of the patients analyzed using tumor-only approaches had genetic alterations in their tumors that were also present in their normal cells, which suggests the alterations were “false-positive” changes not specific to the tumor.
As personalized medicine is predicated on tailoring treatments to the genetic makeup of a patient’s tumor, the high rate of false-positives uncovered in this study has implications for the accuracy of the approach when it relies on tumor-only sequencing, the researchers said.
“We knew from our pioneering whole-exome analyses of cancer patients that a significant number of the genetic alterations that were thought to be associated with tumors were also present in the inherited germline DNA,” said Siân Jones, PhD, of Personal Genome Diagnostics in Baltimore, Maryland.
“By comparing tumor DNA to DNA from normal tissue, we were able to separate out those genetic alterations that are truly tumor-specific. Accurately identifying tumor-specific alterations is essential to realizing the potential of personalized medicine to achieve better treatment outcomes.”
The researchers identified 382 genetic alterations that were potentially tumor-specific by first detecting all of the genetic changes in patients’ tumors and then eliminating those that were well-known germline alterations.
However, when the remaining alterations were compared to the genomic profiles of the patients’ germline DNA, an average of 249, or 65%, turned out to be false-positive changes that were already present in the normal cells.
The researchers also looked at the alterations in actionable genes, which have been identified as potential targets for approved or investigational cancer therapies. They found that almost half of the tumor samples (48%) had at least one false-positive mutation in an actionable gene.
The use of false-positive findings to guide personalized treatment decisions could result in a substantial number of patients receiving therapies that are not optimized for their cancer, according to the researchers. Therefore, it seems sequencing normal DNA alongside tumor DNA is essential.
In addition to selecting personalized therapies for cancer patients, sequencing normal DNA can increase the overall understanding of cancer, including finding cancer predisposition due to germline genome changes, said Victor Velculescu, MD, PhD, of Johns Hopkins University School of Medicine in Baltimore, Maryland.
In this study, the germline analyses revealed changes in cancer-related genes in 3% of the patients who had no known signs of genetically linked cancer.
“These analyses can help us find alterations in cancer-predisposing genes in ways that weren’t previously appreciated,” Dr Velculescu said.
He also acknowledged, however, that there are challenges to implementing tumor-normal DNA analyses in a clinical setting. These include the additional work and costs of sequencing and analyzing a patient’s normal tissue along with his or her tumor tissue.
Costs for tumor gene sequencing begin at several thousand dollars, which would increase if sequencing was done on DNA from normal tissue as well. And health insurance may not fully cover normal-tissue genetic sequencing. ![]()

Image courtesy of NIGMS
Many of the genetic alterations revealed by sequencing a cancer patient’s tumor DNA are not actually associated with the cancer, according to a study published in Science Translational Medicine.
In fact, these alterations are inherited germline mutations already present in an individual’s normal cells.
To make this discovery, researchers compared DNA from tumors and normal cells in 815 patients with hematologic and solid tumor malignancies.
Almost half of the patients analyzed using tumor-only approaches had genetic alterations in their tumors that were also present in their normal cells, which suggests the alterations were “false-positive” changes not specific to the tumor.
As personalized medicine is predicated on tailoring treatments to the genetic makeup of a patient’s tumor, the high rate of false-positives uncovered in this study has implications for the accuracy of the approach when it relies on tumor-only sequencing, the researchers said.
“We knew from our pioneering whole-exome analyses of cancer patients that a significant number of the genetic alterations that were thought to be associated with tumors were also present in the inherited germline DNA,” said Siân Jones, PhD, of Personal Genome Diagnostics in Baltimore, Maryland.
“By comparing tumor DNA to DNA from normal tissue, we were able to separate out those genetic alterations that are truly tumor-specific. Accurately identifying tumor-specific alterations is essential to realizing the potential of personalized medicine to achieve better treatment outcomes.”
The researchers identified 382 genetic alterations that were potentially tumor-specific by first detecting all of the genetic changes in patients’ tumors and then eliminating those that were well-known germline alterations.
However, when the remaining alterations were compared to the genomic profiles of the patients’ germline DNA, an average of 249, or 65%, turned out to be false-positive changes that were already present in the normal cells.
The researchers also looked at the alterations in actionable genes, which have been identified as potential targets for approved or investigational cancer therapies. They found that almost half of the tumor samples (48%) had at least one false-positive mutation in an actionable gene.
The use of false-positive findings to guide personalized treatment decisions could result in a substantial number of patients receiving therapies that are not optimized for their cancer, according to the researchers. Therefore, it seems sequencing normal DNA alongside tumor DNA is essential.
In addition to selecting personalized therapies for cancer patients, sequencing normal DNA can increase the overall understanding of cancer, including finding cancer predisposition due to germline genome changes, said Victor Velculescu, MD, PhD, of Johns Hopkins University School of Medicine in Baltimore, Maryland.
In this study, the germline analyses revealed changes in cancer-related genes in 3% of the patients who had no known signs of genetically linked cancer.
“These analyses can help us find alterations in cancer-predisposing genes in ways that weren’t previously appreciated,” Dr Velculescu said.
He also acknowledged, however, that there are challenges to implementing tumor-normal DNA analyses in a clinical setting. These include the additional work and costs of sequencing and analyzing a patient’s normal tissue along with his or her tumor tissue.
Costs for tumor gene sequencing begin at several thousand dollars, which would increase if sequencing was done on DNA from normal tissue as well. And health insurance may not fully cover normal-tissue genetic sequencing. ![]()
Sleep disorders in patients with cancer
Sleep disturbances are common among patients with cancer for many reasons. Sleep problems can be present at any stage during treatment for cancer and in some patients, sleep disturbance may be the presenting symptoms that lead to the diagnosis of some types of cancer. Poor sleep impairs quality of life In people with cancer, but most do not specifically complain of sleep problems unless they are explicitly asked. Insomnia and fatigue are most common sleep disorders in this cohort, although primary sleep disorders, including obstructive sleep apnea and restless legs syndrome, which are common in the general population, have not been carefully studied in the oncology setting despite significant their impairment of quality of life.
Click on the PDF icon at the top of this introduction to read the full article.
disorder
Sleep disturbances are common among patients with cancer for many reasons. Sleep problems can be present at any stage during treatment for cancer and in some patients, sleep disturbance may be the presenting symptoms that lead to the diagnosis of some types of cancer. Poor sleep impairs quality of life In people with cancer, but most do not specifically complain of sleep problems unless they are explicitly asked. Insomnia and fatigue are most common sleep disorders in this cohort, although primary sleep disorders, including obstructive sleep apnea and restless legs syndrome, which are common in the general population, have not been carefully studied in the oncology setting despite significant their impairment of quality of life.
Click on the PDF icon at the top of this introduction to read the full article.
Sleep disturbances are common among patients with cancer for many reasons. Sleep problems can be present at any stage during treatment for cancer and in some patients, sleep disturbance may be the presenting symptoms that lead to the diagnosis of some types of cancer. Poor sleep impairs quality of life In people with cancer, but most do not specifically complain of sleep problems unless they are explicitly asked. Insomnia and fatigue are most common sleep disorders in this cohort, although primary sleep disorders, including obstructive sleep apnea and restless legs syndrome, which are common in the general population, have not been carefully studied in the oncology setting despite significant their impairment of quality of life.
Click on the PDF icon at the top of this introduction to read the full article.
disorder
disorder
Combo improves PFS in untreated CLL
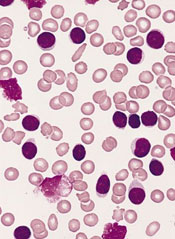
Results of a phase 3 study suggest that adding ofatumumab to chlorambucil can improve progression-free survival (PFS) in treatment-naïve patients with chronic lymphocytic leukemia (CLL).
Ofatumumab plus chlorambucil improved the median PFS by 71% compared to chlorambucil alone.
The combination also improved the overall response rate, duration of response, and time to next treatment.
However, patients in the combination arm had a higher rate of grade 3 or greater adverse events (AEs).
Researchers reported these results in The Lancet. The study, known as COMPLEMENT 1, was funded by GlaxoSmithKline and Genmab A/S.
The study included 447 patients with previously untreated CLL for whom fludarabine-based therapy was considered inappropriate. Patients were randomized to treatment with up to 12 cycles of ofatumumab in combination with chlorambucil (n=221) or up to 12 cycles of chlorambucil alone (n=226).
The study’s primary endpoint was the median PFS, which was 22.4 months in the combination arm and 13.1 months in the chlorambucil arm (hazard ratio [HR]=0.57, P<0.0001). This improvement in PFS was observed in most subgroups, irrespective of age, gender, disease stage, and prognostic factors.
As for secondary endpoints, patients in the combination arm had a higher overall response rate than patients in the chlorambucil arm—82% and 69%, respectively (odds ratio=2.16, P=0.001).
And combination treatment increased the duration of response compared to chlorambucil alone—22.1 months and 13.2 months, respectively (HR=0.56, P<0.001).
Patients in the combination arm also experienced a significantly longer time to next therapy compared to the chlorambucil arm—39.8 months and 24.7 months, respectively (HR=0.49, P<0.0001).
Safety data
The most common AEs (occurring in at least 2% of patients) were neutropenia, thrombocytopenia, anemia, infections, and infusion-related reactions.
Neutropenia occurred more frequently in the combination arm (27% vs 18%), as did infusion-related reactions (67% vs 0%) and infections (46% vs 42%). But thrombocytopenia and anemia were more frequent in the chlorambucil arm (26% vs 14% and 13% vs 9%, respectively).
The incidence of grade 3 or greater AEs was higher in the combination arm than the chlorambucil arm—50% and 43%, respectively.
Grade 3/4 infusion-related reactions occurred in 10% of patients in the combination arm, leading to drug withdrawal in 3% of patients and hospitalization in 2% of patients. No fatal infusion-related reactions were reported.
The most common infections were respiratory tract infections, with an incidence of 27% in the combination arm and 31% in the chlorambucil arm. There were similar frequencies of sepsis (3% and 2%, respectively) and opportunistic infections between the arms (4% and 5%, respectively).
The incidence of AEs leading to treatment withdrawal was 13% in both arms. And the incidence of death during treatment or within 60 days after the last dose was 3% in both arms.
These data formed the basis for regulatory approvals of ofatumumab (Arzerra) in the US and European Union, as well as the recent inclusion of ofatumumab plus chlorambucil in the National Comprehensive Cancer Network treatment guidelines. ![]()

Results of a phase 3 study suggest that adding ofatumumab to chlorambucil can improve progression-free survival (PFS) in treatment-naïve patients with chronic lymphocytic leukemia (CLL).
Ofatumumab plus chlorambucil improved the median PFS by 71% compared to chlorambucil alone.
The combination also improved the overall response rate, duration of response, and time to next treatment.
However, patients in the combination arm had a higher rate of grade 3 or greater adverse events (AEs).
Researchers reported these results in The Lancet. The study, known as COMPLEMENT 1, was funded by GlaxoSmithKline and Genmab A/S.
The study included 447 patients with previously untreated CLL for whom fludarabine-based therapy was considered inappropriate. Patients were randomized to treatment with up to 12 cycles of ofatumumab in combination with chlorambucil (n=221) or up to 12 cycles of chlorambucil alone (n=226).
The study’s primary endpoint was the median PFS, which was 22.4 months in the combination arm and 13.1 months in the chlorambucil arm (hazard ratio [HR]=0.57, P<0.0001). This improvement in PFS was observed in most subgroups, irrespective of age, gender, disease stage, and prognostic factors.
As for secondary endpoints, patients in the combination arm had a higher overall response rate than patients in the chlorambucil arm—82% and 69%, respectively (odds ratio=2.16, P=0.001).
And combination treatment increased the duration of response compared to chlorambucil alone—22.1 months and 13.2 months, respectively (HR=0.56, P<0.001).
Patients in the combination arm also experienced a significantly longer time to next therapy compared to the chlorambucil arm—39.8 months and 24.7 months, respectively (HR=0.49, P<0.0001).
Safety data
The most common AEs (occurring in at least 2% of patients) were neutropenia, thrombocytopenia, anemia, infections, and infusion-related reactions.
Neutropenia occurred more frequently in the combination arm (27% vs 18%), as did infusion-related reactions (67% vs 0%) and infections (46% vs 42%). But thrombocytopenia and anemia were more frequent in the chlorambucil arm (26% vs 14% and 13% vs 9%, respectively).
The incidence of grade 3 or greater AEs was higher in the combination arm than the chlorambucil arm—50% and 43%, respectively.
Grade 3/4 infusion-related reactions occurred in 10% of patients in the combination arm, leading to drug withdrawal in 3% of patients and hospitalization in 2% of patients. No fatal infusion-related reactions were reported.
The most common infections were respiratory tract infections, with an incidence of 27% in the combination arm and 31% in the chlorambucil arm. There were similar frequencies of sepsis (3% and 2%, respectively) and opportunistic infections between the arms (4% and 5%, respectively).
The incidence of AEs leading to treatment withdrawal was 13% in both arms. And the incidence of death during treatment or within 60 days after the last dose was 3% in both arms.
These data formed the basis for regulatory approvals of ofatumumab (Arzerra) in the US and European Union, as well as the recent inclusion of ofatumumab plus chlorambucil in the National Comprehensive Cancer Network treatment guidelines. ![]()

Results of a phase 3 study suggest that adding ofatumumab to chlorambucil can improve progression-free survival (PFS) in treatment-naïve patients with chronic lymphocytic leukemia (CLL).
Ofatumumab plus chlorambucil improved the median PFS by 71% compared to chlorambucil alone.
The combination also improved the overall response rate, duration of response, and time to next treatment.
However, patients in the combination arm had a higher rate of grade 3 or greater adverse events (AEs).
Researchers reported these results in The Lancet. The study, known as COMPLEMENT 1, was funded by GlaxoSmithKline and Genmab A/S.
The study included 447 patients with previously untreated CLL for whom fludarabine-based therapy was considered inappropriate. Patients were randomized to treatment with up to 12 cycles of ofatumumab in combination with chlorambucil (n=221) or up to 12 cycles of chlorambucil alone (n=226).
The study’s primary endpoint was the median PFS, which was 22.4 months in the combination arm and 13.1 months in the chlorambucil arm (hazard ratio [HR]=0.57, P<0.0001). This improvement in PFS was observed in most subgroups, irrespective of age, gender, disease stage, and prognostic factors.
As for secondary endpoints, patients in the combination arm had a higher overall response rate than patients in the chlorambucil arm—82% and 69%, respectively (odds ratio=2.16, P=0.001).
And combination treatment increased the duration of response compared to chlorambucil alone—22.1 months and 13.2 months, respectively (HR=0.56, P<0.001).
Patients in the combination arm also experienced a significantly longer time to next therapy compared to the chlorambucil arm—39.8 months and 24.7 months, respectively (HR=0.49, P<0.0001).
Safety data
The most common AEs (occurring in at least 2% of patients) were neutropenia, thrombocytopenia, anemia, infections, and infusion-related reactions.
Neutropenia occurred more frequently in the combination arm (27% vs 18%), as did infusion-related reactions (67% vs 0%) and infections (46% vs 42%). But thrombocytopenia and anemia were more frequent in the chlorambucil arm (26% vs 14% and 13% vs 9%, respectively).
The incidence of grade 3 or greater AEs was higher in the combination arm than the chlorambucil arm—50% and 43%, respectively.
Grade 3/4 infusion-related reactions occurred in 10% of patients in the combination arm, leading to drug withdrawal in 3% of patients and hospitalization in 2% of patients. No fatal infusion-related reactions were reported.
The most common infections were respiratory tract infections, with an incidence of 27% in the combination arm and 31% in the chlorambucil arm. There were similar frequencies of sepsis (3% and 2%, respectively) and opportunistic infections between the arms (4% and 5%, respectively).
The incidence of AEs leading to treatment withdrawal was 13% in both arms. And the incidence of death during treatment or within 60 days after the last dose was 3% in both arms.
These data formed the basis for regulatory approvals of ofatumumab (Arzerra) in the US and European Union, as well as the recent inclusion of ofatumumab plus chlorambucil in the National Comprehensive Cancer Network treatment guidelines. ![]()
Study reveals how ATRA fights APL

Image courtesy of AFIP
New research suggests the vitamin A derivative all-trans retinoic acid (ATRA) inhibits multiple oncogenic pathways and, at the same time, eliminates cancer stem cells by degrading the Pin1 enzyme.
Investigators said this discovery explains how ATRA successfully treats acute promyelocytic leukemia (APL), and it likely has implications for the treatment of other aggressive or drug-resistant cancers.
The team detailed their discovery in Nature Medicine.
“Pin1 changes protein shape through proline-directed phosphorylation, which is a major control mechanism for disease,” said study author Kun Ping Lu, MD, PhD, of Beth Israel Deaconess Medical Center at Harvard Medical School in Boston, Massachusetts.
“Pin1 is a common, key regulator in many types of cancer and, as a result, can control over 50 oncogenes and tumor suppressors, many of which are known to also control cancer stem cells.”
Until now, agents that inhibit Pin1 have been developed mainly through rational drug design. These inhibitors have proven active against Pin1 in the test tube, but, when tested in a cell model or in vivo, they are unable to efficiently enter cells to successfully inhibit Pin1 function.
In this new work, the investigators decided to take a different approach to identify Pin1 inhibitors. They developed a mechanism-based, high-throughput screen to identify compounds that were targeting active Pin1.
“We had previously identified Pin1 substrate-mimicking peptide inhibitors,” said Xiao Zhen Zhou, MD, also of Beth Israel Deaconess Medical Center.
“We therefore used these as a probe in a competition binding assay and screened approximately 8200 chemical compounds, including both approved drugs and other known bioactive compounds.”
To increase screening success, the investigators chose a probe that specifically binds to the Pin1 enzyme active site very tightly, an approach that is not commonly used for this kind of screen.
“Initially, it appeared that the screening results had no positive hits, so we had to manually sift through them looking for the one that would bind to Pin1,” Dr Zhou said. “We eventually spotted cis retinoic acid, which has the same chemical formula as all-trans retinoic acid but with a different chemical structure.”
It turned out that Pin1 prefers binding to ATRA, and cis retinoic acid needs to convert to ATRA in order to bind Pin1.
ATRA in APL and other cancers
ATRA was first discovered for the treatment of APL in 1987. It was originally thought that ATRA was treating APL by inducing cell differentiation, causing cancer cells to change into normal cells by activating the cellular retinoic acid receptors.
But these new findings suggest that is not the mechanism that is actually behind ATRA’s successful outcomes in treating APL.
“While it has been previously shown that ATRA’s ability to degrade the leukemia-causing fusion oncogene PML-RAR causes ATRA to stop the leukemia stem cells that drive APL, the underlying mechanism has remained elusive,” Dr Lu said.
“Our new, high-throughput drug screening has revealed the ATRA drug target, unexpectedly showing that ATRA directly binds, inhibits, and ultimately degrades active Pin1 selectively in cancer cells. The Pin1-ATRA complex structure suggests that ATRA is trapped in the Pin1 active site by mimicking an unreleasable enzyme substrate. Importantly, ATRA-induced Pin1 ablation degrades the fusion oncogene PML-RAR and treats APL in cell and animal models as well as in human patients.”
The investigators discovered that ATRA-induced Pin1 ablation inhibits triple-negative breast cancer growth as well. The drug proved active in human cells and in animal models, simultaneously turning off many oncogenes and turning on many tumor suppressors.
The team said these results provide a rationale for trying to extend ATRA’s half-life and for developing more potent, Pin1-targeted ATRA variants for cancer treatment.
“The current ATRA drug has a very short half-life of only 45 minutes in humans,” Dr Lu said. “We think that a more potent Pin1 inhibitor will be able to target many ‘dream targets’ that are not currently druggable.”
“ATRA appears to be well tolerated, with minimal side effects, and offers a promising new approach for targeting a Pin1-dependent, common oncogenic mechanism in numerous cancer-driving pathways in cancer and cancer stem cells. This is especially critical for treating aggressive or drug-resistant cancers.” ![]()

Image courtesy of AFIP
New research suggests the vitamin A derivative all-trans retinoic acid (ATRA) inhibits multiple oncogenic pathways and, at the same time, eliminates cancer stem cells by degrading the Pin1 enzyme.
Investigators said this discovery explains how ATRA successfully treats acute promyelocytic leukemia (APL), and it likely has implications for the treatment of other aggressive or drug-resistant cancers.
The team detailed their discovery in Nature Medicine.
“Pin1 changes protein shape through proline-directed phosphorylation, which is a major control mechanism for disease,” said study author Kun Ping Lu, MD, PhD, of Beth Israel Deaconess Medical Center at Harvard Medical School in Boston, Massachusetts.
“Pin1 is a common, key regulator in many types of cancer and, as a result, can control over 50 oncogenes and tumor suppressors, many of which are known to also control cancer stem cells.”
Until now, agents that inhibit Pin1 have been developed mainly through rational drug design. These inhibitors have proven active against Pin1 in the test tube, but, when tested in a cell model or in vivo, they are unable to efficiently enter cells to successfully inhibit Pin1 function.
In this new work, the investigators decided to take a different approach to identify Pin1 inhibitors. They developed a mechanism-based, high-throughput screen to identify compounds that were targeting active Pin1.
“We had previously identified Pin1 substrate-mimicking peptide inhibitors,” said Xiao Zhen Zhou, MD, also of Beth Israel Deaconess Medical Center.
“We therefore used these as a probe in a competition binding assay and screened approximately 8200 chemical compounds, including both approved drugs and other known bioactive compounds.”
To increase screening success, the investigators chose a probe that specifically binds to the Pin1 enzyme active site very tightly, an approach that is not commonly used for this kind of screen.
“Initially, it appeared that the screening results had no positive hits, so we had to manually sift through them looking for the one that would bind to Pin1,” Dr Zhou said. “We eventually spotted cis retinoic acid, which has the same chemical formula as all-trans retinoic acid but with a different chemical structure.”
It turned out that Pin1 prefers binding to ATRA, and cis retinoic acid needs to convert to ATRA in order to bind Pin1.
ATRA in APL and other cancers
ATRA was first discovered for the treatment of APL in 1987. It was originally thought that ATRA was treating APL by inducing cell differentiation, causing cancer cells to change into normal cells by activating the cellular retinoic acid receptors.
But these new findings suggest that is not the mechanism that is actually behind ATRA’s successful outcomes in treating APL.
“While it has been previously shown that ATRA’s ability to degrade the leukemia-causing fusion oncogene PML-RAR causes ATRA to stop the leukemia stem cells that drive APL, the underlying mechanism has remained elusive,” Dr Lu said.
“Our new, high-throughput drug screening has revealed the ATRA drug target, unexpectedly showing that ATRA directly binds, inhibits, and ultimately degrades active Pin1 selectively in cancer cells. The Pin1-ATRA complex structure suggests that ATRA is trapped in the Pin1 active site by mimicking an unreleasable enzyme substrate. Importantly, ATRA-induced Pin1 ablation degrades the fusion oncogene PML-RAR and treats APL in cell and animal models as well as in human patients.”
The investigators discovered that ATRA-induced Pin1 ablation inhibits triple-negative breast cancer growth as well. The drug proved active in human cells and in animal models, simultaneously turning off many oncogenes and turning on many tumor suppressors.
The team said these results provide a rationale for trying to extend ATRA’s half-life and for developing more potent, Pin1-targeted ATRA variants for cancer treatment.
“The current ATRA drug has a very short half-life of only 45 minutes in humans,” Dr Lu said. “We think that a more potent Pin1 inhibitor will be able to target many ‘dream targets’ that are not currently druggable.”
“ATRA appears to be well tolerated, with minimal side effects, and offers a promising new approach for targeting a Pin1-dependent, common oncogenic mechanism in numerous cancer-driving pathways in cancer and cancer stem cells. This is especially critical for treating aggressive or drug-resistant cancers.” ![]()

Image courtesy of AFIP
New research suggests the vitamin A derivative all-trans retinoic acid (ATRA) inhibits multiple oncogenic pathways and, at the same time, eliminates cancer stem cells by degrading the Pin1 enzyme.
Investigators said this discovery explains how ATRA successfully treats acute promyelocytic leukemia (APL), and it likely has implications for the treatment of other aggressive or drug-resistant cancers.
The team detailed their discovery in Nature Medicine.
“Pin1 changes protein shape through proline-directed phosphorylation, which is a major control mechanism for disease,” said study author Kun Ping Lu, MD, PhD, of Beth Israel Deaconess Medical Center at Harvard Medical School in Boston, Massachusetts.
“Pin1 is a common, key regulator in many types of cancer and, as a result, can control over 50 oncogenes and tumor suppressors, many of which are known to also control cancer stem cells.”
Until now, agents that inhibit Pin1 have been developed mainly through rational drug design. These inhibitors have proven active against Pin1 in the test tube, but, when tested in a cell model or in vivo, they are unable to efficiently enter cells to successfully inhibit Pin1 function.
In this new work, the investigators decided to take a different approach to identify Pin1 inhibitors. They developed a mechanism-based, high-throughput screen to identify compounds that were targeting active Pin1.
“We had previously identified Pin1 substrate-mimicking peptide inhibitors,” said Xiao Zhen Zhou, MD, also of Beth Israel Deaconess Medical Center.
“We therefore used these as a probe in a competition binding assay and screened approximately 8200 chemical compounds, including both approved drugs and other known bioactive compounds.”
To increase screening success, the investigators chose a probe that specifically binds to the Pin1 enzyme active site very tightly, an approach that is not commonly used for this kind of screen.
“Initially, it appeared that the screening results had no positive hits, so we had to manually sift through them looking for the one that would bind to Pin1,” Dr Zhou said. “We eventually spotted cis retinoic acid, which has the same chemical formula as all-trans retinoic acid but with a different chemical structure.”
It turned out that Pin1 prefers binding to ATRA, and cis retinoic acid needs to convert to ATRA in order to bind Pin1.
ATRA in APL and other cancers
ATRA was first discovered for the treatment of APL in 1987. It was originally thought that ATRA was treating APL by inducing cell differentiation, causing cancer cells to change into normal cells by activating the cellular retinoic acid receptors.
But these new findings suggest that is not the mechanism that is actually behind ATRA’s successful outcomes in treating APL.
“While it has been previously shown that ATRA’s ability to degrade the leukemia-causing fusion oncogene PML-RAR causes ATRA to stop the leukemia stem cells that drive APL, the underlying mechanism has remained elusive,” Dr Lu said.
“Our new, high-throughput drug screening has revealed the ATRA drug target, unexpectedly showing that ATRA directly binds, inhibits, and ultimately degrades active Pin1 selectively in cancer cells. The Pin1-ATRA complex structure suggests that ATRA is trapped in the Pin1 active site by mimicking an unreleasable enzyme substrate. Importantly, ATRA-induced Pin1 ablation degrades the fusion oncogene PML-RAR and treats APL in cell and animal models as well as in human patients.”
The investigators discovered that ATRA-induced Pin1 ablation inhibits triple-negative breast cancer growth as well. The drug proved active in human cells and in animal models, simultaneously turning off many oncogenes and turning on many tumor suppressors.
The team said these results provide a rationale for trying to extend ATRA’s half-life and for developing more potent, Pin1-targeted ATRA variants for cancer treatment.
“The current ATRA drug has a very short half-life of only 45 minutes in humans,” Dr Lu said. “We think that a more potent Pin1 inhibitor will be able to target many ‘dream targets’ that are not currently druggable.”
“ATRA appears to be well tolerated, with minimal side effects, and offers a promising new approach for targeting a Pin1-dependent, common oncogenic mechanism in numerous cancer-driving pathways in cancer and cancer stem cells. This is especially critical for treating aggressive or drug-resistant cancers.” ![]()
Gene appears key to HSC regulation

in the bone marrow
The gene Ash1l plays a key role in regulating the maintenance and self-renewal of hematopoietic stem cells (HSCs), according to a study published in The Journal of Clinical Investigation.
The research provides new insight into how the body creates and maintains a healthy blood supply and immune system. It also opens new lines of inquiry about Ash1l’s potential role in cancers—like leukemia—that involve other members of the same gene family.
“If we find that Ash1l plays a role [in leukemia], that would open up avenues to try to block or slow down its activity pharmacologically,” said study author Ivan Maillard, MD, of the University of Michigan Medical School in Ann Arbor.
The Ash1l gene regulates the expression of multiple downstream homeotic genes, which help ensure the correct anatomical structure of a developing organism. And Ash1l is part of a family of genes that includes MLL1.
The researchers found that both Ash1l and MLL1 contribute to blood renewal. They observed mild defects in mice missing one gene or the other, but lacking both genes led to catastrophic deficiencies.
“We now have clear evidence that these genes cooperate to develop a healthy blood system,” Dr Maillard said.
He and his colleagues also found that Ash1l-deficient mice had normal numbers of HSCs during early development but a lack of HSCs in maturity—an indication the cells were not able to properly maintain themselves in the bone marrow.
Ash1l-deficient HSCs were unable to establish normal blood renewal after an HSC transplant. Moreover, Ash1l-deficient stem cells competed poorly with normal HSCs in the bone marrow and could easily be dislodged.
“By continuing to investigate the basic, underlying mechanisms [of blood renewal], we are helping to untangle the complex machinery . . . that may lay the foundation for new human treatments 5, 10, or 20 years from now,” Dr Maillard said. ![]()

in the bone marrow
The gene Ash1l plays a key role in regulating the maintenance and self-renewal of hematopoietic stem cells (HSCs), according to a study published in The Journal of Clinical Investigation.
The research provides new insight into how the body creates and maintains a healthy blood supply and immune system. It also opens new lines of inquiry about Ash1l’s potential role in cancers—like leukemia—that involve other members of the same gene family.
“If we find that Ash1l plays a role [in leukemia], that would open up avenues to try to block or slow down its activity pharmacologically,” said study author Ivan Maillard, MD, of the University of Michigan Medical School in Ann Arbor.
The Ash1l gene regulates the expression of multiple downstream homeotic genes, which help ensure the correct anatomical structure of a developing organism. And Ash1l is part of a family of genes that includes MLL1.
The researchers found that both Ash1l and MLL1 contribute to blood renewal. They observed mild defects in mice missing one gene or the other, but lacking both genes led to catastrophic deficiencies.
“We now have clear evidence that these genes cooperate to develop a healthy blood system,” Dr Maillard said.
He and his colleagues also found that Ash1l-deficient mice had normal numbers of HSCs during early development but a lack of HSCs in maturity—an indication the cells were not able to properly maintain themselves in the bone marrow.
Ash1l-deficient HSCs were unable to establish normal blood renewal after an HSC transplant. Moreover, Ash1l-deficient stem cells competed poorly with normal HSCs in the bone marrow and could easily be dislodged.
“By continuing to investigate the basic, underlying mechanisms [of blood renewal], we are helping to untangle the complex machinery . . . that may lay the foundation for new human treatments 5, 10, or 20 years from now,” Dr Maillard said. ![]()

in the bone marrow
The gene Ash1l plays a key role in regulating the maintenance and self-renewal of hematopoietic stem cells (HSCs), according to a study published in The Journal of Clinical Investigation.
The research provides new insight into how the body creates and maintains a healthy blood supply and immune system. It also opens new lines of inquiry about Ash1l’s potential role in cancers—like leukemia—that involve other members of the same gene family.
“If we find that Ash1l plays a role [in leukemia], that would open up avenues to try to block or slow down its activity pharmacologically,” said study author Ivan Maillard, MD, of the University of Michigan Medical School in Ann Arbor.
The Ash1l gene regulates the expression of multiple downstream homeotic genes, which help ensure the correct anatomical structure of a developing organism. And Ash1l is part of a family of genes that includes MLL1.
The researchers found that both Ash1l and MLL1 contribute to blood renewal. They observed mild defects in mice missing one gene or the other, but lacking both genes led to catastrophic deficiencies.
“We now have clear evidence that these genes cooperate to develop a healthy blood system,” Dr Maillard said.
He and his colleagues also found that Ash1l-deficient mice had normal numbers of HSCs during early development but a lack of HSCs in maturity—an indication the cells were not able to properly maintain themselves in the bone marrow.
Ash1l-deficient HSCs were unable to establish normal blood renewal after an HSC transplant. Moreover, Ash1l-deficient stem cells competed poorly with normal HSCs in the bone marrow and could easily be dislodged.
“By continuing to investigate the basic, underlying mechanisms [of blood renewal], we are helping to untangle the complex machinery . . . that may lay the foundation for new human treatments 5, 10, or 20 years from now,” Dr Maillard said. ![]()
Team discovers transmissible leukemia in clams

Photo by Michael J. Metzger
Results of a new study indicate that leukemia may be contagious—at least in clams.
Researchers found that outbreaks of leukemia in soft-shell clams along the east coast of North America can be explained by the spread of cancerous cells from one clam to another.
“The evidence indicates that the tumor cells themselves are contagious—that the cells can spread from one animal to another in the ocean,” said Stephen Goff, PhD, of Columbia University in New York, New York.
“We know this must be true because the genotypes of the tumor cells do not match those of the host animals that acquire the disease, but instead all derive from a single lineage of tumor cells.”
In other words, a leukemia that has killed many clams can be traced to one incidence of disease. The cancer originated in some unfortunate clam and has persisted ever since, as those cancerous cells divide, break free, and make their way to other clams.
Dr Goff and his colleagues described this discovery in Cell.
The researchers noted that only 2 other examples of transmissible cancer are known in the wild. These include the canine transmissible venereal tumor, transmitted by sexual contact, and the Tasmanian devil facial tumor disease, transmitted through biting.
In early studies of the leukemia in clams, Dr Goff and his colleagues found high levels of a particular sequence of DNA they named “Steamer.” While normal cells contain only 2 to 5 copies of Steamer, leukemic cells can have 150 copies. The researchers initially thought this difference was the result of a genetic amplification process occurring within each individual clam.
But when the team analyzed the genomes of leukemia cells collected in New York, Maine, and Prince Edward Island, they discovered something else entirely. The cancerous cells they had collected from clams living at different locations were nearly identical to one another at the genetic level.
“We were astonished to realize that the tumors did not arise from the cells of their diseased host animals, but rather from a rogue clonal cell line spreading over huge geographical distances,” Dr Goff said.
The results showed the cells can survive in seawater long enough to reach and sicken a new host. It is not yet known whether this leukemia can spread to other molluscs or whether there are mechanisms that recognize the malignant cells as foreign invaders and attack them.
Dr Goff said there is plenty the researchers don’t know about this leukemia, including when it first arose and how it spreads from one clam to another. They don’t know what role Steamer played in the cancer’s origin, if any. And they don’t know how often these sorts of cancers might arise in molluscs or other marine animals.
But the findings do suggest that transmissible cancers are more common than anyone suspected. ![]()

Photo by Michael J. Metzger
Results of a new study indicate that leukemia may be contagious—at least in clams.
Researchers found that outbreaks of leukemia in soft-shell clams along the east coast of North America can be explained by the spread of cancerous cells from one clam to another.
“The evidence indicates that the tumor cells themselves are contagious—that the cells can spread from one animal to another in the ocean,” said Stephen Goff, PhD, of Columbia University in New York, New York.
“We know this must be true because the genotypes of the tumor cells do not match those of the host animals that acquire the disease, but instead all derive from a single lineage of tumor cells.”
In other words, a leukemia that has killed many clams can be traced to one incidence of disease. The cancer originated in some unfortunate clam and has persisted ever since, as those cancerous cells divide, break free, and make their way to other clams.
Dr Goff and his colleagues described this discovery in Cell.
The researchers noted that only 2 other examples of transmissible cancer are known in the wild. These include the canine transmissible venereal tumor, transmitted by sexual contact, and the Tasmanian devil facial tumor disease, transmitted through biting.
In early studies of the leukemia in clams, Dr Goff and his colleagues found high levels of a particular sequence of DNA they named “Steamer.” While normal cells contain only 2 to 5 copies of Steamer, leukemic cells can have 150 copies. The researchers initially thought this difference was the result of a genetic amplification process occurring within each individual clam.
But when the team analyzed the genomes of leukemia cells collected in New York, Maine, and Prince Edward Island, they discovered something else entirely. The cancerous cells they had collected from clams living at different locations were nearly identical to one another at the genetic level.
“We were astonished to realize that the tumors did not arise from the cells of their diseased host animals, but rather from a rogue clonal cell line spreading over huge geographical distances,” Dr Goff said.
The results showed the cells can survive in seawater long enough to reach and sicken a new host. It is not yet known whether this leukemia can spread to other molluscs or whether there are mechanisms that recognize the malignant cells as foreign invaders and attack them.
Dr Goff said there is plenty the researchers don’t know about this leukemia, including when it first arose and how it spreads from one clam to another. They don’t know what role Steamer played in the cancer’s origin, if any. And they don’t know how often these sorts of cancers might arise in molluscs or other marine animals.
But the findings do suggest that transmissible cancers are more common than anyone suspected. ![]()

Photo by Michael J. Metzger
Results of a new study indicate that leukemia may be contagious—at least in clams.
Researchers found that outbreaks of leukemia in soft-shell clams along the east coast of North America can be explained by the spread of cancerous cells from one clam to another.
“The evidence indicates that the tumor cells themselves are contagious—that the cells can spread from one animal to another in the ocean,” said Stephen Goff, PhD, of Columbia University in New York, New York.
“We know this must be true because the genotypes of the tumor cells do not match those of the host animals that acquire the disease, but instead all derive from a single lineage of tumor cells.”
In other words, a leukemia that has killed many clams can be traced to one incidence of disease. The cancer originated in some unfortunate clam and has persisted ever since, as those cancerous cells divide, break free, and make their way to other clams.
Dr Goff and his colleagues described this discovery in Cell.
The researchers noted that only 2 other examples of transmissible cancer are known in the wild. These include the canine transmissible venereal tumor, transmitted by sexual contact, and the Tasmanian devil facial tumor disease, transmitted through biting.
In early studies of the leukemia in clams, Dr Goff and his colleagues found high levels of a particular sequence of DNA they named “Steamer.” While normal cells contain only 2 to 5 copies of Steamer, leukemic cells can have 150 copies. The researchers initially thought this difference was the result of a genetic amplification process occurring within each individual clam.
But when the team analyzed the genomes of leukemia cells collected in New York, Maine, and Prince Edward Island, they discovered something else entirely. The cancerous cells they had collected from clams living at different locations were nearly identical to one another at the genetic level.
“We were astonished to realize that the tumors did not arise from the cells of their diseased host animals, but rather from a rogue clonal cell line spreading over huge geographical distances,” Dr Goff said.
The results showed the cells can survive in seawater long enough to reach and sicken a new host. It is not yet known whether this leukemia can spread to other molluscs or whether there are mechanisms that recognize the malignant cells as foreign invaders and attack them.
Dr Goff said there is plenty the researchers don’t know about this leukemia, including when it first arose and how it spreads from one clam to another. They don’t know what role Steamer played in the cancer’s origin, if any. And they don’t know how often these sorts of cancers might arise in molluscs or other marine animals.
But the findings do suggest that transmissible cancers are more common than anyone suspected. ![]()
Drug approved to treat CML, ALL in Canada

Photo courtesy of the FDA
Health Canada has approved ponatinib hydrochloride (Iclusig) to treat adults with any phase of chronic myeloid leukemia (CML) or Philadelphia
chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) for whom other tyrosine kinase inhibitor (TKI) therapy is not appropriate, including CML or Ph+ ALL patients with the T315I mutation and those who have exhibited prior TKI resistance or intolerance.
Ponatinib is approved under the Notice of Compliance with Conditions policy based on promising evidence of clinical effectiveness.
Products approved under this policy are intended for the treatment, prevention, or diagnosis of a serious, life-threatening, or severely debilitating illness. The products must have demonstrated promising benefit, be of high quality, and possess an acceptable safety profile based on a benefit/risk assessment.
These products either respond to a serious unmet medical need in Canada or have demonstrated a significant improvement in the benefit/risk profile over existing therapies.
Ponatinib will be made available in Canada through a controlled distribution program. Prescribers who have completed the certification procedure will be able to prescribe the drug. Trained pharmacies will verify the prescriber’s certified status prior to dispensing ponatinib to the patient.
Health Canada’s decision to approve ponatinib was based on 2-year data from the phase 2 PACE trial.
A trial set to begin in mid-2015 will serve as the confirmatory trial for the Health Canada approval. Investigators will evaluate 3 starting doses of ponatinib in patients with refractory, chronic-phase CML who are resistant to at least 2 approved TKIs.
PACE trial
Researchers conducted this trial in patients with CML or Ph+ ALL who were resistant or intolerant to prior TKI therapy, or who had the T315I mutation.
Ponatinib demonstrated anti-leukemic activity in these patients, prompting a major cytogenetic response (MCyR) in 56% of chronic-phase CML patients and in 70% of patients with the T315I mutation. MCyR within the first 12 months of treatment was the primary endpoint for chronic-phase patients.
In patients with advanced disease, 57% of accelerated-phase CML patients and 31% of blast-phase CML patients achieved a major hematologic response (MaHR). MaHR within the first 6 months was the primary endpoint for patients with advanced disease. In patients with Ph+ ALL, 41% achieved MaHR.
Common non-hematologic adverse events included rash (38%), abdominal pain (38%), headache (35%), dry skin (35%), constipation (34%), fatigue (27%), pyrexia (27%), nausea (26%), arthralgia (25%), hypertension (21%), increased lipase (19%), and increased amylase (7%).
Hematologic events of any grade included thrombocytopenia (42%), neutropenia (24%), and anemia (20%). Serious adverse events of arterial thromboembolism, including arterial stenosis, occurred in patients with cardiovascular risk factors.
Extended follow-up data from the PACE trial, collected in 2013, suggested ponatinib can increase the risk of thrombotic events. When these data came to light, officials in the European Union and the US, where ponatinib had already been approved, began to investigate the drug.
Ponatinib was pulled from the US market for a little over 2 months, and trials of the drug were placed on partial hold while the Food and Drug Administration evaluated the drug’s safety. Ponatinib went back on the market in January 2014, with new safety measures in place.
The drug was not pulled from the market in the European Union, but the European Medicine’s Agency released recommendations for safer use of ponatinib. The Committee for Medicinal Products for Human Use reviewed data on ponatinib and decided the drug’s benefits outweigh its risks. ![]()

Photo courtesy of the FDA
Health Canada has approved ponatinib hydrochloride (Iclusig) to treat adults with any phase of chronic myeloid leukemia (CML) or Philadelphia
chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) for whom other tyrosine kinase inhibitor (TKI) therapy is not appropriate, including CML or Ph+ ALL patients with the T315I mutation and those who have exhibited prior TKI resistance or intolerance.
Ponatinib is approved under the Notice of Compliance with Conditions policy based on promising evidence of clinical effectiveness.
Products approved under this policy are intended for the treatment, prevention, or diagnosis of a serious, life-threatening, or severely debilitating illness. The products must have demonstrated promising benefit, be of high quality, and possess an acceptable safety profile based on a benefit/risk assessment.
These products either respond to a serious unmet medical need in Canada or have demonstrated a significant improvement in the benefit/risk profile over existing therapies.
Ponatinib will be made available in Canada through a controlled distribution program. Prescribers who have completed the certification procedure will be able to prescribe the drug. Trained pharmacies will verify the prescriber’s certified status prior to dispensing ponatinib to the patient.
Health Canada’s decision to approve ponatinib was based on 2-year data from the phase 2 PACE trial.
A trial set to begin in mid-2015 will serve as the confirmatory trial for the Health Canada approval. Investigators will evaluate 3 starting doses of ponatinib in patients with refractory, chronic-phase CML who are resistant to at least 2 approved TKIs.
PACE trial
Researchers conducted this trial in patients with CML or Ph+ ALL who were resistant or intolerant to prior TKI therapy, or who had the T315I mutation.
Ponatinib demonstrated anti-leukemic activity in these patients, prompting a major cytogenetic response (MCyR) in 56% of chronic-phase CML patients and in 70% of patients with the T315I mutation. MCyR within the first 12 months of treatment was the primary endpoint for chronic-phase patients.
In patients with advanced disease, 57% of accelerated-phase CML patients and 31% of blast-phase CML patients achieved a major hematologic response (MaHR). MaHR within the first 6 months was the primary endpoint for patients with advanced disease. In patients with Ph+ ALL, 41% achieved MaHR.
Common non-hematologic adverse events included rash (38%), abdominal pain (38%), headache (35%), dry skin (35%), constipation (34%), fatigue (27%), pyrexia (27%), nausea (26%), arthralgia (25%), hypertension (21%), increased lipase (19%), and increased amylase (7%).
Hematologic events of any grade included thrombocytopenia (42%), neutropenia (24%), and anemia (20%). Serious adverse events of arterial thromboembolism, including arterial stenosis, occurred in patients with cardiovascular risk factors.
Extended follow-up data from the PACE trial, collected in 2013, suggested ponatinib can increase the risk of thrombotic events. When these data came to light, officials in the European Union and the US, where ponatinib had already been approved, began to investigate the drug.
Ponatinib was pulled from the US market for a little over 2 months, and trials of the drug were placed on partial hold while the Food and Drug Administration evaluated the drug’s safety. Ponatinib went back on the market in January 2014, with new safety measures in place.
The drug was not pulled from the market in the European Union, but the European Medicine’s Agency released recommendations for safer use of ponatinib. The Committee for Medicinal Products for Human Use reviewed data on ponatinib and decided the drug’s benefits outweigh its risks. ![]()

Photo courtesy of the FDA
Health Canada has approved ponatinib hydrochloride (Iclusig) to treat adults with any phase of chronic myeloid leukemia (CML) or Philadelphia
chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) for whom other tyrosine kinase inhibitor (TKI) therapy is not appropriate, including CML or Ph+ ALL patients with the T315I mutation and those who have exhibited prior TKI resistance or intolerance.
Ponatinib is approved under the Notice of Compliance with Conditions policy based on promising evidence of clinical effectiveness.
Products approved under this policy are intended for the treatment, prevention, or diagnosis of a serious, life-threatening, or severely debilitating illness. The products must have demonstrated promising benefit, be of high quality, and possess an acceptable safety profile based on a benefit/risk assessment.
These products either respond to a serious unmet medical need in Canada or have demonstrated a significant improvement in the benefit/risk profile over existing therapies.
Ponatinib will be made available in Canada through a controlled distribution program. Prescribers who have completed the certification procedure will be able to prescribe the drug. Trained pharmacies will verify the prescriber’s certified status prior to dispensing ponatinib to the patient.
Health Canada’s decision to approve ponatinib was based on 2-year data from the phase 2 PACE trial.
A trial set to begin in mid-2015 will serve as the confirmatory trial for the Health Canada approval. Investigators will evaluate 3 starting doses of ponatinib in patients with refractory, chronic-phase CML who are resistant to at least 2 approved TKIs.
PACE trial
Researchers conducted this trial in patients with CML or Ph+ ALL who were resistant or intolerant to prior TKI therapy, or who had the T315I mutation.
Ponatinib demonstrated anti-leukemic activity in these patients, prompting a major cytogenetic response (MCyR) in 56% of chronic-phase CML patients and in 70% of patients with the T315I mutation. MCyR within the first 12 months of treatment was the primary endpoint for chronic-phase patients.
In patients with advanced disease, 57% of accelerated-phase CML patients and 31% of blast-phase CML patients achieved a major hematologic response (MaHR). MaHR within the first 6 months was the primary endpoint for patients with advanced disease. In patients with Ph+ ALL, 41% achieved MaHR.
Common non-hematologic adverse events included rash (38%), abdominal pain (38%), headache (35%), dry skin (35%), constipation (34%), fatigue (27%), pyrexia (27%), nausea (26%), arthralgia (25%), hypertension (21%), increased lipase (19%), and increased amylase (7%).
Hematologic events of any grade included thrombocytopenia (42%), neutropenia (24%), and anemia (20%). Serious adverse events of arterial thromboembolism, including arterial stenosis, occurred in patients with cardiovascular risk factors.
Extended follow-up data from the PACE trial, collected in 2013, suggested ponatinib can increase the risk of thrombotic events. When these data came to light, officials in the European Union and the US, where ponatinib had already been approved, began to investigate the drug.
Ponatinib was pulled from the US market for a little over 2 months, and trials of the drug were placed on partial hold while the Food and Drug Administration evaluated the drug’s safety. Ponatinib went back on the market in January 2014, with new safety measures in place.
The drug was not pulled from the market in the European Union, but the European Medicine’s Agency released recommendations for safer use of ponatinib. The Committee for Medicinal Products for Human Use reviewed data on ponatinib and decided the drug’s benefits outweigh its risks.
Protein proves essential for hematopoietic recovery

in the bone marrow
New research suggests the cell survival protein MCL-1, a target for a number of new anticancer agents, is essential for hematopoietic recovery.
Investigators found that reducing MCL-1 levels hindered hematopoietic recovery after chemotherapy and radiotherapy caused extensive destruction of mature blood cells.
Reducing MCL-1 also impaired reconstitution of the bone marrow after hematopoietic stem cell transplant (HSCT).
“Our previous research has shown that targeting MCL-1 could be used with great success for treating certain blood cancers,” said Alex Delbridge, PhD, of the Walter and Eliza Hall Institute of Medical Research in Melbourne, Victoria, Australia.
“However, we have now shown that MCL-1 is also critical for emergency recovery of the blood cell system after cancer therapy-induced blood cell loss.”
Dr Delbridge and his colleagues reported these findings in Blood.
Experiments in mice revealed that loss of a single MCL-1 allele, which reduced MCL-1 protein levels, greatly compromised the immune system and hindered red blood cell recovery after treatment with 5-fluorouracil, γ-irradiation, or HSCT.
Further investigation showed that the pro-apoptotic gene PUMA plays a key role in this phenomenon, as MCL-1 inhibits PUMA. In mice, knocking out PUMA alleviated—but did not eliminate—the HSC survival defect caused by deletion of both MCL-1 alleles.
“This exquisite dependency on MCL-1 for emergency blood cell production has important implications for potential cancer treatments involving MCL-1 inhibitors,” Dr Delbridge said.
“If MCL-1 inhibitors are to be used in combination with other cancer therapies, careful monitoring of the blood cell system will be needed,” added Stephanie Grabow, PhD, also of the Walter and Eliza Hall Institute.
“Our institute colleagues are working to evaluate a potential new drug to treat blood cancers by targeting MCL-1. Our findings suggest that MCL-1 inhibitors and chemotherapeutic drugs should not be used simultaneously.”
Dr Delbridge said this research also offers insights that could help improve HSCT.
“Stem cell transplants can be dangerous because, until the blood cell system is functionally restored, patients are vulnerable to infection,” he said. “Our research suggests that increasing levels of MCL-1 or decreasing the activity of opposing proteins could be a viable strategy for speeding up the regeneration process and reducing the risk of infection after stem cell transplantation.”

in the bone marrow
New research suggests the cell survival protein MCL-1, a target for a number of new anticancer agents, is essential for hematopoietic recovery.
Investigators found that reducing MCL-1 levels hindered hematopoietic recovery after chemotherapy and radiotherapy caused extensive destruction of mature blood cells.
Reducing MCL-1 also impaired reconstitution of the bone marrow after hematopoietic stem cell transplant (HSCT).
“Our previous research has shown that targeting MCL-1 could be used with great success for treating certain blood cancers,” said Alex Delbridge, PhD, of the Walter and Eliza Hall Institute of Medical Research in Melbourne, Victoria, Australia.
“However, we have now shown that MCL-1 is also critical for emergency recovery of the blood cell system after cancer therapy-induced blood cell loss.”
Dr Delbridge and his colleagues reported these findings in Blood.
Experiments in mice revealed that loss of a single MCL-1 allele, which reduced MCL-1 protein levels, greatly compromised the immune system and hindered red blood cell recovery after treatment with 5-fluorouracil, γ-irradiation, or HSCT.
Further investigation showed that the pro-apoptotic gene PUMA plays a key role in this phenomenon, as MCL-1 inhibits PUMA. In mice, knocking out PUMA alleviated—but did not eliminate—the HSC survival defect caused by deletion of both MCL-1 alleles.
“This exquisite dependency on MCL-1 for emergency blood cell production has important implications for potential cancer treatments involving MCL-1 inhibitors,” Dr Delbridge said.
“If MCL-1 inhibitors are to be used in combination with other cancer therapies, careful monitoring of the blood cell system will be needed,” added Stephanie Grabow, PhD, also of the Walter and Eliza Hall Institute.
“Our institute colleagues are working to evaluate a potential new drug to treat blood cancers by targeting MCL-1. Our findings suggest that MCL-1 inhibitors and chemotherapeutic drugs should not be used simultaneously.”
Dr Delbridge said this research also offers insights that could help improve HSCT.
“Stem cell transplants can be dangerous because, until the blood cell system is functionally restored, patients are vulnerable to infection,” he said. “Our research suggests that increasing levels of MCL-1 or decreasing the activity of opposing proteins could be a viable strategy for speeding up the regeneration process and reducing the risk of infection after stem cell transplantation.”

in the bone marrow
New research suggests the cell survival protein MCL-1, a target for a number of new anticancer agents, is essential for hematopoietic recovery.
Investigators found that reducing MCL-1 levels hindered hematopoietic recovery after chemotherapy and radiotherapy caused extensive destruction of mature blood cells.
Reducing MCL-1 also impaired reconstitution of the bone marrow after hematopoietic stem cell transplant (HSCT).
“Our previous research has shown that targeting MCL-1 could be used with great success for treating certain blood cancers,” said Alex Delbridge, PhD, of the Walter and Eliza Hall Institute of Medical Research in Melbourne, Victoria, Australia.
“However, we have now shown that MCL-1 is also critical for emergency recovery of the blood cell system after cancer therapy-induced blood cell loss.”
Dr Delbridge and his colleagues reported these findings in Blood.
Experiments in mice revealed that loss of a single MCL-1 allele, which reduced MCL-1 protein levels, greatly compromised the immune system and hindered red blood cell recovery after treatment with 5-fluorouracil, γ-irradiation, or HSCT.
Further investigation showed that the pro-apoptotic gene PUMA plays a key role in this phenomenon, as MCL-1 inhibits PUMA. In mice, knocking out PUMA alleviated—but did not eliminate—the HSC survival defect caused by deletion of both MCL-1 alleles.
“This exquisite dependency on MCL-1 for emergency blood cell production has important implications for potential cancer treatments involving MCL-1 inhibitors,” Dr Delbridge said.
“If MCL-1 inhibitors are to be used in combination with other cancer therapies, careful monitoring of the blood cell system will be needed,” added Stephanie Grabow, PhD, also of the Walter and Eliza Hall Institute.
“Our institute colleagues are working to evaluate a potential new drug to treat blood cancers by targeting MCL-1. Our findings suggest that MCL-1 inhibitors and chemotherapeutic drugs should not be used simultaneously.”
Dr Delbridge said this research also offers insights that could help improve HSCT.
“Stem cell transplants can be dangerous because, until the blood cell system is functionally restored, patients are vulnerable to infection,” he said. “Our research suggests that increasing levels of MCL-1 or decreasing the activity of opposing proteins could be a viable strategy for speeding up the regeneration process and reducing the risk of infection after stem cell transplantation.”
AML evolution proves unexpectedly complex
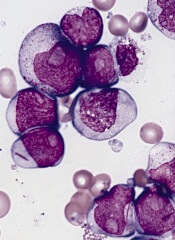
Results of single-cell genotyping suggest the evolution of acute myeloid leukemia (AML) is more complex than we thought.
In screening for mutations in 2 genes, researchers identified at least 9 distinct clonal populations, each harboring unique mutational patterns.
Some mutations seemed to arise not sequentially, but independently and at different points in time.
These results suggest single-cell analysis may be more effective than bulk-tumor analysis for assessing cancer evolution.
Carlo Maley, PhD, of Arizona State University in Tempe, and his colleagues recounted the results in Science Translational Medicine.
The researchers set out to provide a more accurate picture of what takes place at the genetic level when an AML patient experiences relapse or metastasis. So they examined individual cells, screening them for mutations in FLT3 and NPM1.
The results suggested the same mutation was occurring multiple times within an AML patient. The team examined individual cells from 6 AML patients, and the results showed all combinations of homozygous and heterozygous mutations of FLT3 and NPM1.
“There’s no way to explain that with each mutation only happening once,” Dr Maley said. “That’s scary because it means that these cancers have access to many mutations and can find the same mutation over and over.”
Dr Maley noted that the process of convergent evolution, in which separate lineages develop similar features, appears to account for some of the observed diversity. And influences from the environment may drive convergent evolution, but identical mutations can also arise through pure coincidence, simply by virtue of the enormous numbers involved.
For example, a 1 cm3 AML tumor may contain a billion cells, each containing some 3 billion base pairs in its genome. Mutations are estimated to occur at a rate of 1 mutation in every billion base pairs.
“That means every time the population of cells in a 1 cm3 tumor undergoes 1 generation, which we think takes just a couple days, every possible mutation of the genome is happening somewhere in that tumor,” Dr Maley said.
This alone would lead to the same mutation likely occurring independently multiple times.
Curbing cancer’s lethality
Given AML’s near-limitless capacity for creating novel variants, what can clinicians do to halt the disease’s advance? According to Dr Maley, one approach would be to use cancer’s ability to evolve to our advantage, rather than attempt to fight it head on.
This paradigm draws on a branch of ecology known as life history theory. The idea is to carefully study the environmental factors that may lead organisms to favor either a fast or slow reproducing strategy to maximize their ability to survive.
According to the theory, fast reproduction tends to occur in environments with high extrinsic mortality. Aggressive cancer treatment creates just such an environment, favoring those cells able to reproduce quickly, producing large numbers of daughter cells, with a few evading extrinsic mortality to repopulate the tumor.
On the other hand, a very stable environment often favors slow reproduction, because organisms reach a carrying capacity of their surrounding environment. In this case, the limiting factor becomes competition between like organisms. Here, a slow reproducing strategy favoring greater investment in maintenance and survivability wins the competition.
“This approach would say, ‘Let’s keep tumors as stable as possible and keep their resources limited,’” Dr Maley said. “If we are able to keep the tumor cells contained and let them fight it out, we would expect to see more competitively fit cells that are growing very slowly.”

Results of single-cell genotyping suggest the evolution of acute myeloid leukemia (AML) is more complex than we thought.
In screening for mutations in 2 genes, researchers identified at least 9 distinct clonal populations, each harboring unique mutational patterns.
Some mutations seemed to arise not sequentially, but independently and at different points in time.
These results suggest single-cell analysis may be more effective than bulk-tumor analysis for assessing cancer evolution.
Carlo Maley, PhD, of Arizona State University in Tempe, and his colleagues recounted the results in Science Translational Medicine.
The researchers set out to provide a more accurate picture of what takes place at the genetic level when an AML patient experiences relapse or metastasis. So they examined individual cells, screening them for mutations in FLT3 and NPM1.
The results suggested the same mutation was occurring multiple times within an AML patient. The team examined individual cells from 6 AML patients, and the results showed all combinations of homozygous and heterozygous mutations of FLT3 and NPM1.
“There’s no way to explain that with each mutation only happening once,” Dr Maley said. “That’s scary because it means that these cancers have access to many mutations and can find the same mutation over and over.”
Dr Maley noted that the process of convergent evolution, in which separate lineages develop similar features, appears to account for some of the observed diversity. And influences from the environment may drive convergent evolution, but identical mutations can also arise through pure coincidence, simply by virtue of the enormous numbers involved.
For example, a 1 cm3 AML tumor may contain a billion cells, each containing some 3 billion base pairs in its genome. Mutations are estimated to occur at a rate of 1 mutation in every billion base pairs.
“That means every time the population of cells in a 1 cm3 tumor undergoes 1 generation, which we think takes just a couple days, every possible mutation of the genome is happening somewhere in that tumor,” Dr Maley said.
This alone would lead to the same mutation likely occurring independently multiple times.
Curbing cancer’s lethality
Given AML’s near-limitless capacity for creating novel variants, what can clinicians do to halt the disease’s advance? According to Dr Maley, one approach would be to use cancer’s ability to evolve to our advantage, rather than attempt to fight it head on.
This paradigm draws on a branch of ecology known as life history theory. The idea is to carefully study the environmental factors that may lead organisms to favor either a fast or slow reproducing strategy to maximize their ability to survive.
According to the theory, fast reproduction tends to occur in environments with high extrinsic mortality. Aggressive cancer treatment creates just such an environment, favoring those cells able to reproduce quickly, producing large numbers of daughter cells, with a few evading extrinsic mortality to repopulate the tumor.
On the other hand, a very stable environment often favors slow reproduction, because organisms reach a carrying capacity of their surrounding environment. In this case, the limiting factor becomes competition between like organisms. Here, a slow reproducing strategy favoring greater investment in maintenance and survivability wins the competition.
“This approach would say, ‘Let’s keep tumors as stable as possible and keep their resources limited,’” Dr Maley said. “If we are able to keep the tumor cells contained and let them fight it out, we would expect to see more competitively fit cells that are growing very slowly.”

Results of single-cell genotyping suggest the evolution of acute myeloid leukemia (AML) is more complex than we thought.
In screening for mutations in 2 genes, researchers identified at least 9 distinct clonal populations, each harboring unique mutational patterns.
Some mutations seemed to arise not sequentially, but independently and at different points in time.
These results suggest single-cell analysis may be more effective than bulk-tumor analysis for assessing cancer evolution.
Carlo Maley, PhD, of Arizona State University in Tempe, and his colleagues recounted the results in Science Translational Medicine.
The researchers set out to provide a more accurate picture of what takes place at the genetic level when an AML patient experiences relapse or metastasis. So they examined individual cells, screening them for mutations in FLT3 and NPM1.
The results suggested the same mutation was occurring multiple times within an AML patient. The team examined individual cells from 6 AML patients, and the results showed all combinations of homozygous and heterozygous mutations of FLT3 and NPM1.
“There’s no way to explain that with each mutation only happening once,” Dr Maley said. “That’s scary because it means that these cancers have access to many mutations and can find the same mutation over and over.”
Dr Maley noted that the process of convergent evolution, in which separate lineages develop similar features, appears to account for some of the observed diversity. And influences from the environment may drive convergent evolution, but identical mutations can also arise through pure coincidence, simply by virtue of the enormous numbers involved.
For example, a 1 cm3 AML tumor may contain a billion cells, each containing some 3 billion base pairs in its genome. Mutations are estimated to occur at a rate of 1 mutation in every billion base pairs.
“That means every time the population of cells in a 1 cm3 tumor undergoes 1 generation, which we think takes just a couple days, every possible mutation of the genome is happening somewhere in that tumor,” Dr Maley said.
This alone would lead to the same mutation likely occurring independently multiple times.
Curbing cancer’s lethality
Given AML’s near-limitless capacity for creating novel variants, what can clinicians do to halt the disease’s advance? According to Dr Maley, one approach would be to use cancer’s ability to evolve to our advantage, rather than attempt to fight it head on.
This paradigm draws on a branch of ecology known as life history theory. The idea is to carefully study the environmental factors that may lead organisms to favor either a fast or slow reproducing strategy to maximize their ability to survive.
According to the theory, fast reproduction tends to occur in environments with high extrinsic mortality. Aggressive cancer treatment creates just such an environment, favoring those cells able to reproduce quickly, producing large numbers of daughter cells, with a few evading extrinsic mortality to repopulate the tumor.
On the other hand, a very stable environment often favors slow reproduction, because organisms reach a carrying capacity of their surrounding environment. In this case, the limiting factor becomes competition between like organisms. Here, a slow reproducing strategy favoring greater investment in maintenance and survivability wins the competition.
“This approach would say, ‘Let’s keep tumors as stable as possible and keep their resources limited,’” Dr Maley said. “If we are able to keep the tumor cells contained and let them fight it out, we would expect to see more competitively fit cells that are growing very slowly.”