User login
Analyses reveal higher-than-expected pediatric cancer rates in Florida
Photo by Bill Branson
Several statistical analyses have revealed higher-than-expected rates of pediatric cancers in 2 regions of Florida—the Miami metro area and an area west of the Everglades.
The anomalous rates were detected by 5 different research teams—each using different epidemiological and statistical methodology—on a data set spanning the period from 2000 to 2010 that was provided by the Florida Association of Pediatric Tumor Programs (FAPTP).
Lance A. Waller, PhD, of Emory University in Atlanta, Georgia, reviewed the different analyses and described his findings in Statistics and Public Policy.
The research groups applied different analytical approaches to achieve the same goal: detect spatio-temporal pediatric cancer clusters. The analyses used familiar methods—scan statistics, classification, and hierarchical Bayesian modeling—as well as some ideas new to disease clustering: wombling and machine learning.
During their respective analyses of the FAPTP data, the research groups found several suggestive results. For instance, each approach identified local areas in which the observed pediatric cancer rate is significantly higher than the rate expected, given the number of people at risk.
While the precise areas of high reported risk differ between methods, the groups identified a few common results that overlap but are not identical.
For example, all 5 teams identified significantly elevated rates of pediatric cancers in an urban area within collections of ZIP code tabulation areas (ZCTAs) in the Miami metro area and in an area just west of the Everglades. (ZCTAs are geographic areas defined by the US Census Bureau to provide a link between census geography—blocks, block groups, and tracts—and US Postal Service ZIP code areas.)
One analysis suggested the local increase west of the Everglades is based on 2 cases, both classified as “other” race, while another analysis indicated that this cluster is limited to the year 2000.
The observed elevated rates near Miami involved a much larger population size and many more cases, factors that complicate the identification of any shared characteristics common to cases in the cluster.
The analyses also revealed other patterns in the data. Dr Waller said an analysis that revealed a statewide increase in the baseline pediatric cancer incidence rate occurring between 2005 and 2006 merits a closer look to see whether this result represents an overall increase in risk or a change in reporting, because the statistical analysis does not reveal potential reasons for the change.
There also are subtle differences between the specific clusters identified by the various analytical approaches. Comparisons across analyses revealed characteristics of the detected patterns, including the number of cases (2), types of cancer (leukemia or brain/central nervous system cancer), and the racial composition and timing of the cluster west of the Everglades.
As the methods the researchers used don’t completely agree on the precise location, boundaries, and make-up of the detected clusters, the findings suggest a single method may not prove sufficient for such analyses, Dr Waller said.
He added that the identified clusters are geographically quite large and therefore unlikely to provide clear links between particular environmental exposures to local risks.
“While the results do not identify a ‘smoking gun’ in the form of a shared environmental exposure in high-incidence areas, the results do provide epidemiologic insight into the local demographics of the incidence of pediatric cancer cases and suggest more detailed assessment of migration patterns in the Miami area,” Dr Waller said.
“Policy-wise, the results point to responsibly responsive next steps of detailed description of the cases and the at-risk population in the detected areas to summarize local features in the data, particularly the race of cases west of the Everglades and demographic descriptors of any shifts in the at-risk population in the Miami area during the study period.”
Policy responses by local and state health officials may involve more detailed follow-up, including additional data collection, exposure surveys, or in-depth investigation of case histories within a reported cluster.
Dr Waller added that estimated cancer rates consist of the local number of cases (reported by the FAPTP) and the local number of children at risk (reported by the decennial census). Higher-than-expected rates can result from unusually high numbers of reported cases or low numbers of reported local residents.
Since Miami, like many urban areas, often experiences rapid changes in population size and composition between decennial censuses, it is important to assess the accuracy of both data components. Dr Waller suggested, as a first step, assessing the accuracy of the case counts and the inter-census population projections defining the local rates.
“State and local health departments and public health agencies regularly respond to cluster reports from the public,” he said. “Typically, a responsive and effective response is not based on a detailed new epidemiologic study but, rather, is based on education, assessments of local concentrations of demographic risk factors associated with the reported cluster, and an assessment of the distribution of numbers of cases expected given the local demographics.” ![]()

Photo by Bill Branson
Several statistical analyses have revealed higher-than-expected rates of pediatric cancers in 2 regions of Florida—the Miami metro area and an area west of the Everglades.
The anomalous rates were detected by 5 different research teams—each using different epidemiological and statistical methodology—on a data set spanning the period from 2000 to 2010 that was provided by the Florida Association of Pediatric Tumor Programs (FAPTP).
Lance A. Waller, PhD, of Emory University in Atlanta, Georgia, reviewed the different analyses and described his findings in Statistics and Public Policy.
The research groups applied different analytical approaches to achieve the same goal: detect spatio-temporal pediatric cancer clusters. The analyses used familiar methods—scan statistics, classification, and hierarchical Bayesian modeling—as well as some ideas new to disease clustering: wombling and machine learning.
During their respective analyses of the FAPTP data, the research groups found several suggestive results. For instance, each approach identified local areas in which the observed pediatric cancer rate is significantly higher than the rate expected, given the number of people at risk.
While the precise areas of high reported risk differ between methods, the groups identified a few common results that overlap but are not identical.
For example, all 5 teams identified significantly elevated rates of pediatric cancers in an urban area within collections of ZIP code tabulation areas (ZCTAs) in the Miami metro area and in an area just west of the Everglades. (ZCTAs are geographic areas defined by the US Census Bureau to provide a link between census geography—blocks, block groups, and tracts—and US Postal Service ZIP code areas.)
One analysis suggested the local increase west of the Everglades is based on 2 cases, both classified as “other” race, while another analysis indicated that this cluster is limited to the year 2000.
The observed elevated rates near Miami involved a much larger population size and many more cases, factors that complicate the identification of any shared characteristics common to cases in the cluster.
The analyses also revealed other patterns in the data. Dr Waller said an analysis that revealed a statewide increase in the baseline pediatric cancer incidence rate occurring between 2005 and 2006 merits a closer look to see whether this result represents an overall increase in risk or a change in reporting, because the statistical analysis does not reveal potential reasons for the change.
There also are subtle differences between the specific clusters identified by the various analytical approaches. Comparisons across analyses revealed characteristics of the detected patterns, including the number of cases (2), types of cancer (leukemia or brain/central nervous system cancer), and the racial composition and timing of the cluster west of the Everglades.
As the methods the researchers used don’t completely agree on the precise location, boundaries, and make-up of the detected clusters, the findings suggest a single method may not prove sufficient for such analyses, Dr Waller said.
He added that the identified clusters are geographically quite large and therefore unlikely to provide clear links between particular environmental exposures to local risks.
“While the results do not identify a ‘smoking gun’ in the form of a shared environmental exposure in high-incidence areas, the results do provide epidemiologic insight into the local demographics of the incidence of pediatric cancer cases and suggest more detailed assessment of migration patterns in the Miami area,” Dr Waller said.
“Policy-wise, the results point to responsibly responsive next steps of detailed description of the cases and the at-risk population in the detected areas to summarize local features in the data, particularly the race of cases west of the Everglades and demographic descriptors of any shifts in the at-risk population in the Miami area during the study period.”
Policy responses by local and state health officials may involve more detailed follow-up, including additional data collection, exposure surveys, or in-depth investigation of case histories within a reported cluster.
Dr Waller added that estimated cancer rates consist of the local number of cases (reported by the FAPTP) and the local number of children at risk (reported by the decennial census). Higher-than-expected rates can result from unusually high numbers of reported cases or low numbers of reported local residents.
Since Miami, like many urban areas, often experiences rapid changes in population size and composition between decennial censuses, it is important to assess the accuracy of both data components. Dr Waller suggested, as a first step, assessing the accuracy of the case counts and the inter-census population projections defining the local rates.
“State and local health departments and public health agencies regularly respond to cluster reports from the public,” he said. “Typically, a responsive and effective response is not based on a detailed new epidemiologic study but, rather, is based on education, assessments of local concentrations of demographic risk factors associated with the reported cluster, and an assessment of the distribution of numbers of cases expected given the local demographics.” ![]()

Photo by Bill Branson
Several statistical analyses have revealed higher-than-expected rates of pediatric cancers in 2 regions of Florida—the Miami metro area and an area west of the Everglades.
The anomalous rates were detected by 5 different research teams—each using different epidemiological and statistical methodology—on a data set spanning the period from 2000 to 2010 that was provided by the Florida Association of Pediatric Tumor Programs (FAPTP).
Lance A. Waller, PhD, of Emory University in Atlanta, Georgia, reviewed the different analyses and described his findings in Statistics and Public Policy.
The research groups applied different analytical approaches to achieve the same goal: detect spatio-temporal pediatric cancer clusters. The analyses used familiar methods—scan statistics, classification, and hierarchical Bayesian modeling—as well as some ideas new to disease clustering: wombling and machine learning.
During their respective analyses of the FAPTP data, the research groups found several suggestive results. For instance, each approach identified local areas in which the observed pediatric cancer rate is significantly higher than the rate expected, given the number of people at risk.
While the precise areas of high reported risk differ between methods, the groups identified a few common results that overlap but are not identical.
For example, all 5 teams identified significantly elevated rates of pediatric cancers in an urban area within collections of ZIP code tabulation areas (ZCTAs) in the Miami metro area and in an area just west of the Everglades. (ZCTAs are geographic areas defined by the US Census Bureau to provide a link between census geography—blocks, block groups, and tracts—and US Postal Service ZIP code areas.)
One analysis suggested the local increase west of the Everglades is based on 2 cases, both classified as “other” race, while another analysis indicated that this cluster is limited to the year 2000.
The observed elevated rates near Miami involved a much larger population size and many more cases, factors that complicate the identification of any shared characteristics common to cases in the cluster.
The analyses also revealed other patterns in the data. Dr Waller said an analysis that revealed a statewide increase in the baseline pediatric cancer incidence rate occurring between 2005 and 2006 merits a closer look to see whether this result represents an overall increase in risk or a change in reporting, because the statistical analysis does not reveal potential reasons for the change.
There also are subtle differences between the specific clusters identified by the various analytical approaches. Comparisons across analyses revealed characteristics of the detected patterns, including the number of cases (2), types of cancer (leukemia or brain/central nervous system cancer), and the racial composition and timing of the cluster west of the Everglades.
As the methods the researchers used don’t completely agree on the precise location, boundaries, and make-up of the detected clusters, the findings suggest a single method may not prove sufficient for such analyses, Dr Waller said.
He added that the identified clusters are geographically quite large and therefore unlikely to provide clear links between particular environmental exposures to local risks.
“While the results do not identify a ‘smoking gun’ in the form of a shared environmental exposure in high-incidence areas, the results do provide epidemiologic insight into the local demographics of the incidence of pediatric cancer cases and suggest more detailed assessment of migration patterns in the Miami area,” Dr Waller said.
“Policy-wise, the results point to responsibly responsive next steps of detailed description of the cases and the at-risk population in the detected areas to summarize local features in the data, particularly the race of cases west of the Everglades and demographic descriptors of any shifts in the at-risk population in the Miami area during the study period.”
Policy responses by local and state health officials may involve more detailed follow-up, including additional data collection, exposure surveys, or in-depth investigation of case histories within a reported cluster.
Dr Waller added that estimated cancer rates consist of the local number of cases (reported by the FAPTP) and the local number of children at risk (reported by the decennial census). Higher-than-expected rates can result from unusually high numbers of reported cases or low numbers of reported local residents.
Since Miami, like many urban areas, often experiences rapid changes in population size and composition between decennial censuses, it is important to assess the accuracy of both data components. Dr Waller suggested, as a first step, assessing the accuracy of the case counts and the inter-census population projections defining the local rates.
“State and local health departments and public health agencies regularly respond to cluster reports from the public,” he said. “Typically, a responsive and effective response is not based on a detailed new epidemiologic study but, rather, is based on education, assessments of local concentrations of demographic risk factors associated with the reported cluster, and an assessment of the distribution of numbers of cases expected given the local demographics.” ![]()
ABVD and Stanford V similar for bulky mediastinal Hodgkin’s lymphoma
Failure-free survival and overall survival were similar between two combined modality therapies in patients with stage I or II bulky mediastinal Hodgkin’s lymphoma, investigators reported.
The results were published online April 20 in the Journal of Clinical Oncology.
The phase III trial evaluated outcomes following treatment with either doxorubicin, bleomycin, vinblastine, and dacarbazine (ABVD) or mechlorethamine, doxorubicin, vincristine, bleomycin, vinblastine, etoposide, and prednisone (Stanford V).
Median failure-free survival (FFS) and overall survival (OS) were not reached in either arm. The 5-year FFS and OS were 85% and 96% for ABVD, respectively, and 79% and 92% for Stanford V, reported Dr. Ranjana H. Advani, professor of oncology at Stanford (Calif.) University, and associates.
At a median follow up of 6.54 years, 19 treatment failures occurred in the ABVD arm and 23 in the Stanford V arm. In total, 14 deaths occurred, 5 in the ABVD group and 9 in the Stanford V group.
Approximately 20%-25% of patients with stage I or II Hodgkin’s lymphoma (HL) have bulky mediastinal involvement, and this was the first contemporary prospective trial to evaluate this patient subgroup, the investigators wrote (J. Clin. Oncol. 2015 April 20 [doi:10.1200/JCO.2014.57.8138]).
“This is important because ongoing trials in North America use mediastinal bulk as an eligibility criterion, and contemporary guidelines use it to define treatment algorithms,” Dr. Advani and associates said, noting that both regimens are acceptable treatment options.
“In addition, these results provide an important contemporary benchmark for comparison of ongoing and future studies,” they wrote.
Out of 854 patients with HL enrolled in the trial, 264 with bulky disease were eligible for the subgroup analysis; 135 received ABVD and 129 received Stanford V. After completion of chemotherapy, all patients received 36 Gy of modified involved field radiotherapy (IFRT). Patterns of relapse were similar between treatment arms, and less than 10% of patients had in-field recurrences, a finding that indicated effective local control with IFRT.
Both treatment arms had similar rates of grade 3-4 neutropenia, and the Stanford V arm had more grade 3 lymphopenia (83% vs. 46%, P < .001) and grade 3 and 4 sensory neuropathy. At 5 years, both groups had similar risks of second cancers: two in the ABVD group and six in the Stanford group. The assessment of risks associated with higher doses of anthracycline and bleomycin in ABVD and larger radiation fields in Stanford V requires longer follow-up, the researchers wrote.
Failure-free survival and overall survival were similar between two combined modality therapies in patients with stage I or II bulky mediastinal Hodgkin’s lymphoma, investigators reported.
The results were published online April 20 in the Journal of Clinical Oncology.
The phase III trial evaluated outcomes following treatment with either doxorubicin, bleomycin, vinblastine, and dacarbazine (ABVD) or mechlorethamine, doxorubicin, vincristine, bleomycin, vinblastine, etoposide, and prednisone (Stanford V).
Median failure-free survival (FFS) and overall survival (OS) were not reached in either arm. The 5-year FFS and OS were 85% and 96% for ABVD, respectively, and 79% and 92% for Stanford V, reported Dr. Ranjana H. Advani, professor of oncology at Stanford (Calif.) University, and associates.
At a median follow up of 6.54 years, 19 treatment failures occurred in the ABVD arm and 23 in the Stanford V arm. In total, 14 deaths occurred, 5 in the ABVD group and 9 in the Stanford V group.
Approximately 20%-25% of patients with stage I or II Hodgkin’s lymphoma (HL) have bulky mediastinal involvement, and this was the first contemporary prospective trial to evaluate this patient subgroup, the investigators wrote (J. Clin. Oncol. 2015 April 20 [doi:10.1200/JCO.2014.57.8138]).
“This is important because ongoing trials in North America use mediastinal bulk as an eligibility criterion, and contemporary guidelines use it to define treatment algorithms,” Dr. Advani and associates said, noting that both regimens are acceptable treatment options.
“In addition, these results provide an important contemporary benchmark for comparison of ongoing and future studies,” they wrote.
Out of 854 patients with HL enrolled in the trial, 264 with bulky disease were eligible for the subgroup analysis; 135 received ABVD and 129 received Stanford V. After completion of chemotherapy, all patients received 36 Gy of modified involved field radiotherapy (IFRT). Patterns of relapse were similar between treatment arms, and less than 10% of patients had in-field recurrences, a finding that indicated effective local control with IFRT.
Both treatment arms had similar rates of grade 3-4 neutropenia, and the Stanford V arm had more grade 3 lymphopenia (83% vs. 46%, P < .001) and grade 3 and 4 sensory neuropathy. At 5 years, both groups had similar risks of second cancers: two in the ABVD group and six in the Stanford group. The assessment of risks associated with higher doses of anthracycline and bleomycin in ABVD and larger radiation fields in Stanford V requires longer follow-up, the researchers wrote.
Failure-free survival and overall survival were similar between two combined modality therapies in patients with stage I or II bulky mediastinal Hodgkin’s lymphoma, investigators reported.
The results were published online April 20 in the Journal of Clinical Oncology.
The phase III trial evaluated outcomes following treatment with either doxorubicin, bleomycin, vinblastine, and dacarbazine (ABVD) or mechlorethamine, doxorubicin, vincristine, bleomycin, vinblastine, etoposide, and prednisone (Stanford V).
Median failure-free survival (FFS) and overall survival (OS) were not reached in either arm. The 5-year FFS and OS were 85% and 96% for ABVD, respectively, and 79% and 92% for Stanford V, reported Dr. Ranjana H. Advani, professor of oncology at Stanford (Calif.) University, and associates.
At a median follow up of 6.54 years, 19 treatment failures occurred in the ABVD arm and 23 in the Stanford V arm. In total, 14 deaths occurred, 5 in the ABVD group and 9 in the Stanford V group.
Approximately 20%-25% of patients with stage I or II Hodgkin’s lymphoma (HL) have bulky mediastinal involvement, and this was the first contemporary prospective trial to evaluate this patient subgroup, the investigators wrote (J. Clin. Oncol. 2015 April 20 [doi:10.1200/JCO.2014.57.8138]).
“This is important because ongoing trials in North America use mediastinal bulk as an eligibility criterion, and contemporary guidelines use it to define treatment algorithms,” Dr. Advani and associates said, noting that both regimens are acceptable treatment options.
“In addition, these results provide an important contemporary benchmark for comparison of ongoing and future studies,” they wrote.
Out of 854 patients with HL enrolled in the trial, 264 with bulky disease were eligible for the subgroup analysis; 135 received ABVD and 129 received Stanford V. After completion of chemotherapy, all patients received 36 Gy of modified involved field radiotherapy (IFRT). Patterns of relapse were similar between treatment arms, and less than 10% of patients had in-field recurrences, a finding that indicated effective local control with IFRT.
Both treatment arms had similar rates of grade 3-4 neutropenia, and the Stanford V arm had more grade 3 lymphopenia (83% vs. 46%, P < .001) and grade 3 and 4 sensory neuropathy. At 5 years, both groups had similar risks of second cancers: two in the ABVD group and six in the Stanford group. The assessment of risks associated with higher doses of anthracycline and bleomycin in ABVD and larger radiation fields in Stanford V requires longer follow-up, the researchers wrote.
FROM JOURNAL OF CLINICAL ONCOLOGY
Key clinical point: No significant differences in outcomes were observed after treatment with ABVD vs. Stanford V in patients with stage I or II bulky mediastinal Hodgkin’s lymphoma.
Major finding: At a median follow-up of 6.5 years, ABVD and Stanford V resulted in similar numbers of treatment failures (19 vs. 23), complete remission rate (75% vs. 81%), and overall response rate (83% vs. 88%), respectively.
Data source: A subgroup analysis of a phase III trial of patients with stage I or II bulky disease, in which 135 were assigned ABVD and 129 received Stanford V.
Disclosures: Dr. Advani reported receiving research funds from Millennium, Takeda Oncology, Seattle Genetics, Genentech/Roche, Allos Therapeutics, Pharmacyclics, Janssen Pharmaceuticals, Celgene, and Idera Pharmaceuticals. Many of his coauthors reported ties to several industry sources.
EBV-CTLs produce durable responses in EBV-LPD
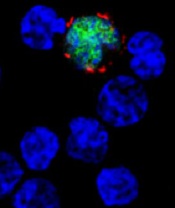
among uninfected cells (blue)
Image courtesy of NIH/
Benjamin Chaigne-Delalande
PHILADELPHIA—Cytotoxic T lymphocytes designed to target Epstein-Barr virus (EBV-CTLs) can elicit durable responses in patients
with EBV–associated lymphoproliferative disorder (EBV-LPD), according to data presented at the AACRAnnual Meeting 2015.
Results from two trials showed that EBV-CTLs derived from a patient’s transplant donor could produce a response rate of 62%, and EBV-CTLs derived from third-party donors could produce a response rate of 61%.
Study investigators noted that, with the achievement of complete response (CR), remission proved durable. And, unlike with chemotherapy, partial responses (PRs) to EBV-CTLs were durable as well.
The team presented these results as abstract CT107.*
“The purpose of our clinical trials was to see if giving T cells from a normal-immune individual that were expanded in culture and stimulated to respond to multiple proteins from the Epstein-Barr virus could provide a safe and effective treatment,” said Richard J. O’Reilly, MD, of Memorial Sloan Kettering Cancer Center in New York.
“The good news from our two clinical trials is that EBV-CTLs generated from either the patient’s transplant donor or from the bank of normal donor T cells developed at Memorial Sloan Kettering put aggressive EBV-LPD that had failed to respond to rituximab into long-lasting remission in more than 60% of patients.”
In the first trial, 26 patients with EBV-LPD received EBV-CTLs generated from their transplant donor. Thirteen of these patients had previously received rituximab, and 16 had high-risk disease.
Thirteen patients in this trial received HLA-matched, EBV-CTLs from the Memorial Sloan Kettering Cancer Center bank of EBV-CTLs generated from third-party, healthy donors. All 13 patients had high-risk disease, and 12 had received prior rituximab.
Dr O’Reilly noted that good results were observed with EBV-CTLs from both sources in this trial. And because EBV-CTLs from the bank are available immediately, he and his team used only EBV-CTLs from the bank when treating the 18 patients enrolled in the second trial.
Among the 39 patients enrolled in the first trial, 23 had a CR, none had a PR, and 2 had stable disease.
For patients who received EBV-CTLs from their primary donor, the combined rate of CR and PR was 62% (16 CRs). For patients who received third-party EBV-CTLs, the combined rate of CR and PR was 54% (7 CRs).
Sixteen of the patients who achieved a CR are still doing well, Dr O’Reilly said. Eight of these patients are alive more than 5 years after receiving EBV-CTLs, and 1 is alive more than 10 years after treatment.
Among the 18 patients enrolled in the second trial, 9 had a CR, 3 had a PR, and 1 had stable disease. The combined rate of CR and PR was 67%.
All of the patients who achieved a CR in this trial continue to do well, and the investigators will be following them long-term, Dr O’Reilly said.
He also noted that toxicities with EBV-CTLs were minimal, and there were no treatment-related deaths. None of the patients developed cytokine release syndrome or graft-vs-host disease requiring systemic therapy.
“The EBV-CTLs work well for the majority of recipients,” Dr O’Reilly said. “However, the responses became clinically evident only after the T cells expanded in vivo, which took about 7 to 14 days. We are rigorously pursuing the development of biomarkers or other tests to predict response earlier.”
Memorial Sloan Kettering Cancer Center has entered into an option agreement with Atara Biotherapeutics to further develop EBV-CTLs for clinical use. However, the data presented at AACR were accrued prior to that agreement.
Last month, the US Food and Drug Administration granted breakthrough therapy designation to EBV-CTLs generated from third-party donors for the treatment of patients with rituximab-refractory EBV-LPD. ![]()
*Information in the abstract differs from that presented at the meeting.

among uninfected cells (blue)
Image courtesy of NIH/
Benjamin Chaigne-Delalande
PHILADELPHIA—Cytotoxic T lymphocytes designed to target Epstein-Barr virus (EBV-CTLs) can elicit durable responses in patients
with EBV–associated lymphoproliferative disorder (EBV-LPD), according to data presented at the AACRAnnual Meeting 2015.
Results from two trials showed that EBV-CTLs derived from a patient’s transplant donor could produce a response rate of 62%, and EBV-CTLs derived from third-party donors could produce a response rate of 61%.
Study investigators noted that, with the achievement of complete response (CR), remission proved durable. And, unlike with chemotherapy, partial responses (PRs) to EBV-CTLs were durable as well.
The team presented these results as abstract CT107.*
“The purpose of our clinical trials was to see if giving T cells from a normal-immune individual that were expanded in culture and stimulated to respond to multiple proteins from the Epstein-Barr virus could provide a safe and effective treatment,” said Richard J. O’Reilly, MD, of Memorial Sloan Kettering Cancer Center in New York.
“The good news from our two clinical trials is that EBV-CTLs generated from either the patient’s transplant donor or from the bank of normal donor T cells developed at Memorial Sloan Kettering put aggressive EBV-LPD that had failed to respond to rituximab into long-lasting remission in more than 60% of patients.”
In the first trial, 26 patients with EBV-LPD received EBV-CTLs generated from their transplant donor. Thirteen of these patients had previously received rituximab, and 16 had high-risk disease.
Thirteen patients in this trial received HLA-matched, EBV-CTLs from the Memorial Sloan Kettering Cancer Center bank of EBV-CTLs generated from third-party, healthy donors. All 13 patients had high-risk disease, and 12 had received prior rituximab.
Dr O’Reilly noted that good results were observed with EBV-CTLs from both sources in this trial. And because EBV-CTLs from the bank are available immediately, he and his team used only EBV-CTLs from the bank when treating the 18 patients enrolled in the second trial.
Among the 39 patients enrolled in the first trial, 23 had a CR, none had a PR, and 2 had stable disease.
For patients who received EBV-CTLs from their primary donor, the combined rate of CR and PR was 62% (16 CRs). For patients who received third-party EBV-CTLs, the combined rate of CR and PR was 54% (7 CRs).
Sixteen of the patients who achieved a CR are still doing well, Dr O’Reilly said. Eight of these patients are alive more than 5 years after receiving EBV-CTLs, and 1 is alive more than 10 years after treatment.
Among the 18 patients enrolled in the second trial, 9 had a CR, 3 had a PR, and 1 had stable disease. The combined rate of CR and PR was 67%.
All of the patients who achieved a CR in this trial continue to do well, and the investigators will be following them long-term, Dr O’Reilly said.
He also noted that toxicities with EBV-CTLs were minimal, and there were no treatment-related deaths. None of the patients developed cytokine release syndrome or graft-vs-host disease requiring systemic therapy.
“The EBV-CTLs work well for the majority of recipients,” Dr O’Reilly said. “However, the responses became clinically evident only after the T cells expanded in vivo, which took about 7 to 14 days. We are rigorously pursuing the development of biomarkers or other tests to predict response earlier.”
Memorial Sloan Kettering Cancer Center has entered into an option agreement with Atara Biotherapeutics to further develop EBV-CTLs for clinical use. However, the data presented at AACR were accrued prior to that agreement.
Last month, the US Food and Drug Administration granted breakthrough therapy designation to EBV-CTLs generated from third-party donors for the treatment of patients with rituximab-refractory EBV-LPD. ![]()
*Information in the abstract differs from that presented at the meeting.

among uninfected cells (blue)
Image courtesy of NIH/
Benjamin Chaigne-Delalande
PHILADELPHIA—Cytotoxic T lymphocytes designed to target Epstein-Barr virus (EBV-CTLs) can elicit durable responses in patients
with EBV–associated lymphoproliferative disorder (EBV-LPD), according to data presented at the AACRAnnual Meeting 2015.
Results from two trials showed that EBV-CTLs derived from a patient’s transplant donor could produce a response rate of 62%, and EBV-CTLs derived from third-party donors could produce a response rate of 61%.
Study investigators noted that, with the achievement of complete response (CR), remission proved durable. And, unlike with chemotherapy, partial responses (PRs) to EBV-CTLs were durable as well.
The team presented these results as abstract CT107.*
“The purpose of our clinical trials was to see if giving T cells from a normal-immune individual that were expanded in culture and stimulated to respond to multiple proteins from the Epstein-Barr virus could provide a safe and effective treatment,” said Richard J. O’Reilly, MD, of Memorial Sloan Kettering Cancer Center in New York.
“The good news from our two clinical trials is that EBV-CTLs generated from either the patient’s transplant donor or from the bank of normal donor T cells developed at Memorial Sloan Kettering put aggressive EBV-LPD that had failed to respond to rituximab into long-lasting remission in more than 60% of patients.”
In the first trial, 26 patients with EBV-LPD received EBV-CTLs generated from their transplant donor. Thirteen of these patients had previously received rituximab, and 16 had high-risk disease.
Thirteen patients in this trial received HLA-matched, EBV-CTLs from the Memorial Sloan Kettering Cancer Center bank of EBV-CTLs generated from third-party, healthy donors. All 13 patients had high-risk disease, and 12 had received prior rituximab.
Dr O’Reilly noted that good results were observed with EBV-CTLs from both sources in this trial. And because EBV-CTLs from the bank are available immediately, he and his team used only EBV-CTLs from the bank when treating the 18 patients enrolled in the second trial.
Among the 39 patients enrolled in the first trial, 23 had a CR, none had a PR, and 2 had stable disease.
For patients who received EBV-CTLs from their primary donor, the combined rate of CR and PR was 62% (16 CRs). For patients who received third-party EBV-CTLs, the combined rate of CR and PR was 54% (7 CRs).
Sixteen of the patients who achieved a CR are still doing well, Dr O’Reilly said. Eight of these patients are alive more than 5 years after receiving EBV-CTLs, and 1 is alive more than 10 years after treatment.
Among the 18 patients enrolled in the second trial, 9 had a CR, 3 had a PR, and 1 had stable disease. The combined rate of CR and PR was 67%.
All of the patients who achieved a CR in this trial continue to do well, and the investigators will be following them long-term, Dr O’Reilly said.
He also noted that toxicities with EBV-CTLs were minimal, and there were no treatment-related deaths. None of the patients developed cytokine release syndrome or graft-vs-host disease requiring systemic therapy.
“The EBV-CTLs work well for the majority of recipients,” Dr O’Reilly said. “However, the responses became clinically evident only after the T cells expanded in vivo, which took about 7 to 14 days. We are rigorously pursuing the development of biomarkers or other tests to predict response earlier.”
Memorial Sloan Kettering Cancer Center has entered into an option agreement with Atara Biotherapeutics to further develop EBV-CTLs for clinical use. However, the data presented at AACR were accrued prior to that agreement.
Last month, the US Food and Drug Administration granted breakthrough therapy designation to EBV-CTLs generated from third-party donors for the treatment of patients with rituximab-refractory EBV-LPD. ![]()
*Information in the abstract differs from that presented at the meeting.
New mAb can overcome resistance to other mAbs

Photo courtesy of the
University of Southampton
A newly developed monoclonal antibody (mAb) can reverse resistance to other mAbs in chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL), according to research published in Cancer Cell.
Investigators found that some cancer cells draw mAbs inside themselves, making them invisible to immune cells.
But a mAb called BI-1206 can prevent this process and enhance cancer killing by binding to a molecule called FcγRIIB.
In preclinical experiments, BI-1206 was able to overcome resistance to mAbs such as rituximab.
“With more monoclonal antibody treatments being developed, there is an urgent need to understand how tumors become resistant to them and develop ways to overcome it,” said study author Mark Cragg, PhD, of the University of Southampton in the UK.
“Not only does BI-1206 appear to be able to reverse resistance to a range of monoclonal antibodies, it is also effective at directly killing cancer cells itself.”
In the Cancer Cell paper, BI-1206 is referred to as 6G11. The investigators found that 6G11 can block rituximab internalization and has “potent antitumor activity” in vitro. 6G11 was also well-tolerated and did not prompt cytokine storm.
In a mouse model of CLL, 6G11 enhanced rituximab-mediated depletion of primary CLL cells and improved responses when compared to rituximab alone.
In mice engrafted with cells from patients with CLL that was refractory to rituximab, ofatumumab, and/or alemtuzumab, 6G11 alone depleted CLL cells but did not improve overall response rates compared to rituximab alone. However, 6G11 in combination with rituximab did improve overall response rates compared to rituximab alone.
In a mouse model of MCL, neither 6G11 nor rituximab alone improved long-term survival. However, 30% of mice treated with both drugs survived tumor-free out to 100 days.
Combining 6G11 with obinutuzumab significantly improved splenic tumor cell depletion in mice with CLL. And more than 90% of mice that received 6G11 and alemtuzumab had a complete response to the treatment.
The investigators said these data suggest 6G11 can overcome mAb resistance for multiple targets. They said the drug will be tested in patients with CLL and non-Hodgkin lymphoma in an early stage clinical trial. ![]()

Photo courtesy of the
University of Southampton
A newly developed monoclonal antibody (mAb) can reverse resistance to other mAbs in chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL), according to research published in Cancer Cell.
Investigators found that some cancer cells draw mAbs inside themselves, making them invisible to immune cells.
But a mAb called BI-1206 can prevent this process and enhance cancer killing by binding to a molecule called FcγRIIB.
In preclinical experiments, BI-1206 was able to overcome resistance to mAbs such as rituximab.
“With more monoclonal antibody treatments being developed, there is an urgent need to understand how tumors become resistant to them and develop ways to overcome it,” said study author Mark Cragg, PhD, of the University of Southampton in the UK.
“Not only does BI-1206 appear to be able to reverse resistance to a range of monoclonal antibodies, it is also effective at directly killing cancer cells itself.”
In the Cancer Cell paper, BI-1206 is referred to as 6G11. The investigators found that 6G11 can block rituximab internalization and has “potent antitumor activity” in vitro. 6G11 was also well-tolerated and did not prompt cytokine storm.
In a mouse model of CLL, 6G11 enhanced rituximab-mediated depletion of primary CLL cells and improved responses when compared to rituximab alone.
In mice engrafted with cells from patients with CLL that was refractory to rituximab, ofatumumab, and/or alemtuzumab, 6G11 alone depleted CLL cells but did not improve overall response rates compared to rituximab alone. However, 6G11 in combination with rituximab did improve overall response rates compared to rituximab alone.
In a mouse model of MCL, neither 6G11 nor rituximab alone improved long-term survival. However, 30% of mice treated with both drugs survived tumor-free out to 100 days.
Combining 6G11 with obinutuzumab significantly improved splenic tumor cell depletion in mice with CLL. And more than 90% of mice that received 6G11 and alemtuzumab had a complete response to the treatment.
The investigators said these data suggest 6G11 can overcome mAb resistance for multiple targets. They said the drug will be tested in patients with CLL and non-Hodgkin lymphoma in an early stage clinical trial. ![]()

Photo courtesy of the
University of Southampton
A newly developed monoclonal antibody (mAb) can reverse resistance to other mAbs in chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL), according to research published in Cancer Cell.
Investigators found that some cancer cells draw mAbs inside themselves, making them invisible to immune cells.
But a mAb called BI-1206 can prevent this process and enhance cancer killing by binding to a molecule called FcγRIIB.
In preclinical experiments, BI-1206 was able to overcome resistance to mAbs such as rituximab.
“With more monoclonal antibody treatments being developed, there is an urgent need to understand how tumors become resistant to them and develop ways to overcome it,” said study author Mark Cragg, PhD, of the University of Southampton in the UK.
“Not only does BI-1206 appear to be able to reverse resistance to a range of monoclonal antibodies, it is also effective at directly killing cancer cells itself.”
In the Cancer Cell paper, BI-1206 is referred to as 6G11. The investigators found that 6G11 can block rituximab internalization and has “potent antitumor activity” in vitro. 6G11 was also well-tolerated and did not prompt cytokine storm.
In a mouse model of CLL, 6G11 enhanced rituximab-mediated depletion of primary CLL cells and improved responses when compared to rituximab alone.
In mice engrafted with cells from patients with CLL that was refractory to rituximab, ofatumumab, and/or alemtuzumab, 6G11 alone depleted CLL cells but did not improve overall response rates compared to rituximab alone. However, 6G11 in combination with rituximab did improve overall response rates compared to rituximab alone.
In a mouse model of MCL, neither 6G11 nor rituximab alone improved long-term survival. However, 30% of mice treated with both drugs survived tumor-free out to 100 days.
Combining 6G11 with obinutuzumab significantly improved splenic tumor cell depletion in mice with CLL. And more than 90% of mice that received 6G11 and alemtuzumab had a complete response to the treatment.
The investigators said these data suggest 6G11 can overcome mAb resistance for multiple targets. They said the drug will be tested in patients with CLL and non-Hodgkin lymphoma in an early stage clinical trial. ![]()
Tumor sequencing fails to paint the whole picture

Image courtesy of NIGMS
Many of the genetic alterations revealed by sequencing a cancer patient’s tumor DNA are not actually associated with the cancer, according to a study published in Science Translational Medicine.
In fact, these alterations are inherited germline mutations already present in an individual’s normal cells.
To make this discovery, researchers compared DNA from tumors and normal cells in 815 patients with hematologic and solid tumor malignancies.
Almost half of the patients analyzed using tumor-only approaches had genetic alterations in their tumors that were also present in their normal cells, which suggests the alterations were “false-positive” changes not specific to the tumor.
As personalized medicine is predicated on tailoring treatments to the genetic makeup of a patient’s tumor, the high rate of false-positives uncovered in this study has implications for the accuracy of the approach when it relies on tumor-only sequencing, the researchers said.
“We knew from our pioneering whole-exome analyses of cancer patients that a significant number of the genetic alterations that were thought to be associated with tumors were also present in the inherited germline DNA,” said Siân Jones, PhD, of Personal Genome Diagnostics in Baltimore, Maryland.
“By comparing tumor DNA to DNA from normal tissue, we were able to separate out those genetic alterations that are truly tumor-specific. Accurately identifying tumor-specific alterations is essential to realizing the potential of personalized medicine to achieve better treatment outcomes.”
The researchers identified 382 genetic alterations that were potentially tumor-specific by first detecting all of the genetic changes in patients’ tumors and then eliminating those that were well-known germline alterations.
However, when the remaining alterations were compared to the genomic profiles of the patients’ germline DNA, an average of 249, or 65%, turned out to be false-positive changes that were already present in the normal cells.
The researchers also looked at the alterations in actionable genes, which have been identified as potential targets for approved or investigational cancer therapies. They found that almost half of the tumor samples (48%) had at least one false-positive mutation in an actionable gene.
The use of false-positive findings to guide personalized treatment decisions could result in a substantial number of patients receiving therapies that are not optimized for their cancer, according to the researchers. Therefore, it seems sequencing normal DNA alongside tumor DNA is essential.
In addition to selecting personalized therapies for cancer patients, sequencing normal DNA can increase the overall understanding of cancer, including finding cancer predisposition due to germline genome changes, said Victor Velculescu, MD, PhD, of Johns Hopkins University School of Medicine in Baltimore, Maryland.
In this study, the germline analyses revealed changes in cancer-related genes in 3% of the patients who had no known signs of genetically linked cancer.
“These analyses can help us find alterations in cancer-predisposing genes in ways that weren’t previously appreciated,” Dr Velculescu said.
He also acknowledged, however, that there are challenges to implementing tumor-normal DNA analyses in a clinical setting. These include the additional work and costs of sequencing and analyzing a patient’s normal tissue along with his or her tumor tissue.
Costs for tumor gene sequencing begin at several thousand dollars, which would increase if sequencing was done on DNA from normal tissue as well. And health insurance may not fully cover normal-tissue genetic sequencing. ![]()

Image courtesy of NIGMS
Many of the genetic alterations revealed by sequencing a cancer patient’s tumor DNA are not actually associated with the cancer, according to a study published in Science Translational Medicine.
In fact, these alterations are inherited germline mutations already present in an individual’s normal cells.
To make this discovery, researchers compared DNA from tumors and normal cells in 815 patients with hematologic and solid tumor malignancies.
Almost half of the patients analyzed using tumor-only approaches had genetic alterations in their tumors that were also present in their normal cells, which suggests the alterations were “false-positive” changes not specific to the tumor.
As personalized medicine is predicated on tailoring treatments to the genetic makeup of a patient’s tumor, the high rate of false-positives uncovered in this study has implications for the accuracy of the approach when it relies on tumor-only sequencing, the researchers said.
“We knew from our pioneering whole-exome analyses of cancer patients that a significant number of the genetic alterations that were thought to be associated with tumors were also present in the inherited germline DNA,” said Siân Jones, PhD, of Personal Genome Diagnostics in Baltimore, Maryland.
“By comparing tumor DNA to DNA from normal tissue, we were able to separate out those genetic alterations that are truly tumor-specific. Accurately identifying tumor-specific alterations is essential to realizing the potential of personalized medicine to achieve better treatment outcomes.”
The researchers identified 382 genetic alterations that were potentially tumor-specific by first detecting all of the genetic changes in patients’ tumors and then eliminating those that were well-known germline alterations.
However, when the remaining alterations were compared to the genomic profiles of the patients’ germline DNA, an average of 249, or 65%, turned out to be false-positive changes that were already present in the normal cells.
The researchers also looked at the alterations in actionable genes, which have been identified as potential targets for approved or investigational cancer therapies. They found that almost half of the tumor samples (48%) had at least one false-positive mutation in an actionable gene.
The use of false-positive findings to guide personalized treatment decisions could result in a substantial number of patients receiving therapies that are not optimized for their cancer, according to the researchers. Therefore, it seems sequencing normal DNA alongside tumor DNA is essential.
In addition to selecting personalized therapies for cancer patients, sequencing normal DNA can increase the overall understanding of cancer, including finding cancer predisposition due to germline genome changes, said Victor Velculescu, MD, PhD, of Johns Hopkins University School of Medicine in Baltimore, Maryland.
In this study, the germline analyses revealed changes in cancer-related genes in 3% of the patients who had no known signs of genetically linked cancer.
“These analyses can help us find alterations in cancer-predisposing genes in ways that weren’t previously appreciated,” Dr Velculescu said.
He also acknowledged, however, that there are challenges to implementing tumor-normal DNA analyses in a clinical setting. These include the additional work and costs of sequencing and analyzing a patient’s normal tissue along with his or her tumor tissue.
Costs for tumor gene sequencing begin at several thousand dollars, which would increase if sequencing was done on DNA from normal tissue as well. And health insurance may not fully cover normal-tissue genetic sequencing. ![]()

Image courtesy of NIGMS
Many of the genetic alterations revealed by sequencing a cancer patient’s tumor DNA are not actually associated with the cancer, according to a study published in Science Translational Medicine.
In fact, these alterations are inherited germline mutations already present in an individual’s normal cells.
To make this discovery, researchers compared DNA from tumors and normal cells in 815 patients with hematologic and solid tumor malignancies.
Almost half of the patients analyzed using tumor-only approaches had genetic alterations in their tumors that were also present in their normal cells, which suggests the alterations were “false-positive” changes not specific to the tumor.
As personalized medicine is predicated on tailoring treatments to the genetic makeup of a patient’s tumor, the high rate of false-positives uncovered in this study has implications for the accuracy of the approach when it relies on tumor-only sequencing, the researchers said.
“We knew from our pioneering whole-exome analyses of cancer patients that a significant number of the genetic alterations that were thought to be associated with tumors were also present in the inherited germline DNA,” said Siân Jones, PhD, of Personal Genome Diagnostics in Baltimore, Maryland.
“By comparing tumor DNA to DNA from normal tissue, we were able to separate out those genetic alterations that are truly tumor-specific. Accurately identifying tumor-specific alterations is essential to realizing the potential of personalized medicine to achieve better treatment outcomes.”
The researchers identified 382 genetic alterations that were potentially tumor-specific by first detecting all of the genetic changes in patients’ tumors and then eliminating those that were well-known germline alterations.
However, when the remaining alterations were compared to the genomic profiles of the patients’ germline DNA, an average of 249, or 65%, turned out to be false-positive changes that were already present in the normal cells.
The researchers also looked at the alterations in actionable genes, which have been identified as potential targets for approved or investigational cancer therapies. They found that almost half of the tumor samples (48%) had at least one false-positive mutation in an actionable gene.
The use of false-positive findings to guide personalized treatment decisions could result in a substantial number of patients receiving therapies that are not optimized for their cancer, according to the researchers. Therefore, it seems sequencing normal DNA alongside tumor DNA is essential.
In addition to selecting personalized therapies for cancer patients, sequencing normal DNA can increase the overall understanding of cancer, including finding cancer predisposition due to germline genome changes, said Victor Velculescu, MD, PhD, of Johns Hopkins University School of Medicine in Baltimore, Maryland.
In this study, the germline analyses revealed changes in cancer-related genes in 3% of the patients who had no known signs of genetically linked cancer.
“These analyses can help us find alterations in cancer-predisposing genes in ways that weren’t previously appreciated,” Dr Velculescu said.
He also acknowledged, however, that there are challenges to implementing tumor-normal DNA analyses in a clinical setting. These include the additional work and costs of sequencing and analyzing a patient’s normal tissue along with his or her tumor tissue.
Costs for tumor gene sequencing begin at several thousand dollars, which would increase if sequencing was done on DNA from normal tissue as well. And health insurance may not fully cover normal-tissue genetic sequencing. ![]()
Technique helps determine DLBCL subtypes

Photo by Darren Baker
A new technique can quickly and easily differentiate the two major subtypes of diffuse large B-cell lymphoma (DLBCL), researchers have reported in The Journal of Molecular Diagnostics.
The method is based on a reverse transcriptase multiplex ligation-dependent probe amplification (RT-MLPA) assay and 14 gene signatures.
It proved as accurate as the gold standard technology for identifying germinal center B-cell-like (GCB) and activated B-cell-like (ABC) DLBCL.
So the researchers believe this RT-MLPA-based assay could help clinicians evaluate a patient’s prognosis and choose the optimal therapy for a person with DLBCL.
“Differences in the progression of the disease and clinical outcomes can, at least in part, be explained by the heterogeneity of lymphoma, which can be classified into two major subtypes with different outcomes,” said study author Philippe Ruminy, PhD, of the University of Rouen in France.
“Unfortunately, these lymphomas are morphologically undistinguishable in routine diagnosis, which is a major problem for the development of targeted therapies. Furthermore, array-based gene expression profiling (GEP), which is considered the gold standard for discriminating these tumors, remains poorly transposable to routine diagnosis, and the surrogate immunohistochemical (IHC) algorithms that have been proposed are often considered poorly reliable.”
With this in mind, Dr Ruminy and his colleagues tested the RT-MLPA-based assay alongside GEP techniques in samples from 259 patients with de novo DLBCL.
The team used a series of 195 patients from a single institution to train and validate the new technique. Of these patients, 115 had a classification determined with a Veracode DASL assay, 100 had an IHC classification, and 135 had survival data. A second series of 64 patients that were previously analyzed by Affymetrix U133+2 GEP arrays served as an external validation cohort.
When the researchers looked at 50 patients randomly selected from the training cohort, they found that the RT-MLPA-based assay classified 90% of the cases within the expected subtypes (20 GCB and 25 ABC). The remaining 10% (1 GCB and 4 ABC) were considered unclassified.
In the external validation cohort, the RT-MLPA-based assay classified 80% of cases within the expected subtypes (24 ABC and 28 GCB). However, 13.8% were considered unclassified (3 GCB and 6 ABC), and 4 were misclassified.
The researchers also found the RT-MLPA-based assay was sensitive for analyzing archived formalin-fixed, paraffin-embedded (FFPE) tissue samples. A comparison of samples from paired frozen and FFPE biopsies showed that the technique correctly classified 89.3% of 28 cases.
“Because RT-MLPA requires only short cDNA fragments for the correct binding and ligation of the gene-specific oligonucleotide probes, it is less affected by the use of low RNA concentrations and RNA degradation,” Dr Ruminy said.
“It could thus be used for the retrospective analysis of archival collections and for the inclusion of patients in prospective clinical trials, because only a few institutions routinely collect frozen biopsy material.”
To evaluate the prognostic value of the assay, the researchers looked at survival in 135 treated DLBCL patients who were diagnosed between 2001 and 2011.
Patients determined to have the ABC subtype by the RT-MLPA-based assay had significantly worse progression-free survival and overall survival than patients with the GCB subtype. And the expression of several individual genes within the MLPA signature was significantly associated with prognosis (ie, high LMO2, high BCL6, and low TNFRSF13B expression).
“The robust and cost-effective RT-MLPA assay can yield results within one day and requires reagents costing less than $5 per sample,” Dr Ruminy said. “Since RT-MLPA utilizes materials and equipment that are standard in many laboratories, the process can easily be implemented for routine use.” ![]()

Photo by Darren Baker
A new technique can quickly and easily differentiate the two major subtypes of diffuse large B-cell lymphoma (DLBCL), researchers have reported in The Journal of Molecular Diagnostics.
The method is based on a reverse transcriptase multiplex ligation-dependent probe amplification (RT-MLPA) assay and 14 gene signatures.
It proved as accurate as the gold standard technology for identifying germinal center B-cell-like (GCB) and activated B-cell-like (ABC) DLBCL.
So the researchers believe this RT-MLPA-based assay could help clinicians evaluate a patient’s prognosis and choose the optimal therapy for a person with DLBCL.
“Differences in the progression of the disease and clinical outcomes can, at least in part, be explained by the heterogeneity of lymphoma, which can be classified into two major subtypes with different outcomes,” said study author Philippe Ruminy, PhD, of the University of Rouen in France.
“Unfortunately, these lymphomas are morphologically undistinguishable in routine diagnosis, which is a major problem for the development of targeted therapies. Furthermore, array-based gene expression profiling (GEP), which is considered the gold standard for discriminating these tumors, remains poorly transposable to routine diagnosis, and the surrogate immunohistochemical (IHC) algorithms that have been proposed are often considered poorly reliable.”
With this in mind, Dr Ruminy and his colleagues tested the RT-MLPA-based assay alongside GEP techniques in samples from 259 patients with de novo DLBCL.
The team used a series of 195 patients from a single institution to train and validate the new technique. Of these patients, 115 had a classification determined with a Veracode DASL assay, 100 had an IHC classification, and 135 had survival data. A second series of 64 patients that were previously analyzed by Affymetrix U133+2 GEP arrays served as an external validation cohort.
When the researchers looked at 50 patients randomly selected from the training cohort, they found that the RT-MLPA-based assay classified 90% of the cases within the expected subtypes (20 GCB and 25 ABC). The remaining 10% (1 GCB and 4 ABC) were considered unclassified.
In the external validation cohort, the RT-MLPA-based assay classified 80% of cases within the expected subtypes (24 ABC and 28 GCB). However, 13.8% were considered unclassified (3 GCB and 6 ABC), and 4 were misclassified.
The researchers also found the RT-MLPA-based assay was sensitive for analyzing archived formalin-fixed, paraffin-embedded (FFPE) tissue samples. A comparison of samples from paired frozen and FFPE biopsies showed that the technique correctly classified 89.3% of 28 cases.
“Because RT-MLPA requires only short cDNA fragments for the correct binding and ligation of the gene-specific oligonucleotide probes, it is less affected by the use of low RNA concentrations and RNA degradation,” Dr Ruminy said.
“It could thus be used for the retrospective analysis of archival collections and for the inclusion of patients in prospective clinical trials, because only a few institutions routinely collect frozen biopsy material.”
To evaluate the prognostic value of the assay, the researchers looked at survival in 135 treated DLBCL patients who were diagnosed between 2001 and 2011.
Patients determined to have the ABC subtype by the RT-MLPA-based assay had significantly worse progression-free survival and overall survival than patients with the GCB subtype. And the expression of several individual genes within the MLPA signature was significantly associated with prognosis (ie, high LMO2, high BCL6, and low TNFRSF13B expression).
“The robust and cost-effective RT-MLPA assay can yield results within one day and requires reagents costing less than $5 per sample,” Dr Ruminy said. “Since RT-MLPA utilizes materials and equipment that are standard in many laboratories, the process can easily be implemented for routine use.” ![]()

Photo by Darren Baker
A new technique can quickly and easily differentiate the two major subtypes of diffuse large B-cell lymphoma (DLBCL), researchers have reported in The Journal of Molecular Diagnostics.
The method is based on a reverse transcriptase multiplex ligation-dependent probe amplification (RT-MLPA) assay and 14 gene signatures.
It proved as accurate as the gold standard technology for identifying germinal center B-cell-like (GCB) and activated B-cell-like (ABC) DLBCL.
So the researchers believe this RT-MLPA-based assay could help clinicians evaluate a patient’s prognosis and choose the optimal therapy for a person with DLBCL.
“Differences in the progression of the disease and clinical outcomes can, at least in part, be explained by the heterogeneity of lymphoma, which can be classified into two major subtypes with different outcomes,” said study author Philippe Ruminy, PhD, of the University of Rouen in France.
“Unfortunately, these lymphomas are morphologically undistinguishable in routine diagnosis, which is a major problem for the development of targeted therapies. Furthermore, array-based gene expression profiling (GEP), which is considered the gold standard for discriminating these tumors, remains poorly transposable to routine diagnosis, and the surrogate immunohistochemical (IHC) algorithms that have been proposed are often considered poorly reliable.”
With this in mind, Dr Ruminy and his colleagues tested the RT-MLPA-based assay alongside GEP techniques in samples from 259 patients with de novo DLBCL.
The team used a series of 195 patients from a single institution to train and validate the new technique. Of these patients, 115 had a classification determined with a Veracode DASL assay, 100 had an IHC classification, and 135 had survival data. A second series of 64 patients that were previously analyzed by Affymetrix U133+2 GEP arrays served as an external validation cohort.
When the researchers looked at 50 patients randomly selected from the training cohort, they found that the RT-MLPA-based assay classified 90% of the cases within the expected subtypes (20 GCB and 25 ABC). The remaining 10% (1 GCB and 4 ABC) were considered unclassified.
In the external validation cohort, the RT-MLPA-based assay classified 80% of cases within the expected subtypes (24 ABC and 28 GCB). However, 13.8% were considered unclassified (3 GCB and 6 ABC), and 4 were misclassified.
The researchers also found the RT-MLPA-based assay was sensitive for analyzing archived formalin-fixed, paraffin-embedded (FFPE) tissue samples. A comparison of samples from paired frozen and FFPE biopsies showed that the technique correctly classified 89.3% of 28 cases.
“Because RT-MLPA requires only short cDNA fragments for the correct binding and ligation of the gene-specific oligonucleotide probes, it is less affected by the use of low RNA concentrations and RNA degradation,” Dr Ruminy said.
“It could thus be used for the retrospective analysis of archival collections and for the inclusion of patients in prospective clinical trials, because only a few institutions routinely collect frozen biopsy material.”
To evaluate the prognostic value of the assay, the researchers looked at survival in 135 treated DLBCL patients who were diagnosed between 2001 and 2011.
Patients determined to have the ABC subtype by the RT-MLPA-based assay had significantly worse progression-free survival and overall survival than patients with the GCB subtype. And the expression of several individual genes within the MLPA signature was significantly associated with prognosis (ie, high LMO2, high BCL6, and low TNFRSF13B expression).
“The robust and cost-effective RT-MLPA assay can yield results within one day and requires reagents costing less than $5 per sample,” Dr Ruminy said. “Since RT-MLPA utilizes materials and equipment that are standard in many laboratories, the process can easily be implemented for routine use.” ![]()
Sleep disorders in patients with cancer
Sleep disturbances are common among patients with cancer for many reasons. Sleep problems can be present at any stage during treatment for cancer and in some patients, sleep disturbance may be the presenting symptoms that lead to the diagnosis of some types of cancer. Poor sleep impairs quality of life In people with cancer, but most do not specifically complain of sleep problems unless they are explicitly asked. Insomnia and fatigue are most common sleep disorders in this cohort, although primary sleep disorders, including obstructive sleep apnea and restless legs syndrome, which are common in the general population, have not been carefully studied in the oncology setting despite significant their impairment of quality of life.
Click on the PDF icon at the top of this introduction to read the full article.
disorder
Sleep disturbances are common among patients with cancer for many reasons. Sleep problems can be present at any stage during treatment for cancer and in some patients, sleep disturbance may be the presenting symptoms that lead to the diagnosis of some types of cancer. Poor sleep impairs quality of life In people with cancer, but most do not specifically complain of sleep problems unless they are explicitly asked. Insomnia and fatigue are most common sleep disorders in this cohort, although primary sleep disorders, including obstructive sleep apnea and restless legs syndrome, which are common in the general population, have not been carefully studied in the oncology setting despite significant their impairment of quality of life.
Click on the PDF icon at the top of this introduction to read the full article.
Sleep disturbances are common among patients with cancer for many reasons. Sleep problems can be present at any stage during treatment for cancer and in some patients, sleep disturbance may be the presenting symptoms that lead to the diagnosis of some types of cancer. Poor sleep impairs quality of life In people with cancer, but most do not specifically complain of sleep problems unless they are explicitly asked. Insomnia and fatigue are most common sleep disorders in this cohort, although primary sleep disorders, including obstructive sleep apnea and restless legs syndrome, which are common in the general population, have not been carefully studied in the oncology setting despite significant their impairment of quality of life.
Click on the PDF icon at the top of this introduction to read the full article.
disorder
disorder
A new treatment approach for ALK- ALCL?
Investigators say they have uncovered a new approach for treating ALK-negative anaplastic large cell lymphoma (ALCL).
Results of genomic analyses indicated that many cases of ALK-negative ALCL may be driven by alterations in the JAK/STAT3 pathway, and in vivo
experiments showed the disease can be inhibited by compounds that target this pathway.
The investigators believe these compounds could be more effective than current therapies.
“Current therapies for this form of lymphoma fail to work in the majority of cases,” said Raul Rabadan, PhD, of Columbia University in New York, New York.
“However, now that we know the mutations that drive a significant percentage of cases, we can envision a new, personalized genomic approach to the treatment of ALK-negative ALCL.”
Dr Rabadan and his colleagues recounted their discovery of the mutations in Cancer Cell.
The team had sequenced the exomes and RNA of cancer cells from 155 patients with ALCL and 74 control subjects with other types of lymphoma.
Results revealed mutations in either JAK1 or STAT3 in about 20% of the 88 patients with ALK-negative ALCL. Of that 20%, 38% had mutations in both genes.
The investigators also detected the presence of several novel gene fusions (NFκB2-ROS1, NFκB2-TYK2, NCOC2-ROS1, and PABPC4-TYK2), some of which appear to activate the JAK/STAT3 pathway (NFκB2-ROS1 and NFκB2-TYK2).
Patients with these fusions did not have JAK1 or STAT3 mutations, which suggests the fusions are an independent cause of ALK-negative ALCL.
To confirm whether JAK1 and STAT3 mutations can cause ALK-negative ALCL, the investigators induced these mutations in normal human cells. The mutations did, in fact, lead to diseased cells.
Finally, the team tested JAK/STAT3 pathway inhibitors—ruxolitinib and PUH71—in mouse models of ALK-negative ALCL. And they found that both drugs significantly inhibited tumor growth.
“Our findings demonstrate that drugs targeting the JAK/STAT3 pathway offer a viable therapeutic strategy in a subset of patients with ALCL,” Dr Rabadan said.
“A couple of JAK/STAT3 inhibitors have been approved by the FDA for the treatment of psoriasis and rheumatoid arthritis, and several more are currently in clinical trials. These could be tested in patients whose genetic profile matches those we identified in our study.” ![]()

Investigators say they have uncovered a new approach for treating ALK-negative anaplastic large cell lymphoma (ALCL).
Results of genomic analyses indicated that many cases of ALK-negative ALCL may be driven by alterations in the JAK/STAT3 pathway, and in vivo
experiments showed the disease can be inhibited by compounds that target this pathway.
The investigators believe these compounds could be more effective than current therapies.
“Current therapies for this form of lymphoma fail to work in the majority of cases,” said Raul Rabadan, PhD, of Columbia University in New York, New York.
“However, now that we know the mutations that drive a significant percentage of cases, we can envision a new, personalized genomic approach to the treatment of ALK-negative ALCL.”
Dr Rabadan and his colleagues recounted their discovery of the mutations in Cancer Cell.
The team had sequenced the exomes and RNA of cancer cells from 155 patients with ALCL and 74 control subjects with other types of lymphoma.
Results revealed mutations in either JAK1 or STAT3 in about 20% of the 88 patients with ALK-negative ALCL. Of that 20%, 38% had mutations in both genes.
The investigators also detected the presence of several novel gene fusions (NFκB2-ROS1, NFκB2-TYK2, NCOC2-ROS1, and PABPC4-TYK2), some of which appear to activate the JAK/STAT3 pathway (NFκB2-ROS1 and NFκB2-TYK2).
Patients with these fusions did not have JAK1 or STAT3 mutations, which suggests the fusions are an independent cause of ALK-negative ALCL.
To confirm whether JAK1 and STAT3 mutations can cause ALK-negative ALCL, the investigators induced these mutations in normal human cells. The mutations did, in fact, lead to diseased cells.
Finally, the team tested JAK/STAT3 pathway inhibitors—ruxolitinib and PUH71—in mouse models of ALK-negative ALCL. And they found that both drugs significantly inhibited tumor growth.
“Our findings demonstrate that drugs targeting the JAK/STAT3 pathway offer a viable therapeutic strategy in a subset of patients with ALCL,” Dr Rabadan said.
“A couple of JAK/STAT3 inhibitors have been approved by the FDA for the treatment of psoriasis and rheumatoid arthritis, and several more are currently in clinical trials. These could be tested in patients whose genetic profile matches those we identified in our study.” ![]()

Investigators say they have uncovered a new approach for treating ALK-negative anaplastic large cell lymphoma (ALCL).
Results of genomic analyses indicated that many cases of ALK-negative ALCL may be driven by alterations in the JAK/STAT3 pathway, and in vivo
experiments showed the disease can be inhibited by compounds that target this pathway.
The investigators believe these compounds could be more effective than current therapies.
“Current therapies for this form of lymphoma fail to work in the majority of cases,” said Raul Rabadan, PhD, of Columbia University in New York, New York.
“However, now that we know the mutations that drive a significant percentage of cases, we can envision a new, personalized genomic approach to the treatment of ALK-negative ALCL.”
Dr Rabadan and his colleagues recounted their discovery of the mutations in Cancer Cell.
The team had sequenced the exomes and RNA of cancer cells from 155 patients with ALCL and 74 control subjects with other types of lymphoma.
Results revealed mutations in either JAK1 or STAT3 in about 20% of the 88 patients with ALK-negative ALCL. Of that 20%, 38% had mutations in both genes.
The investigators also detected the presence of several novel gene fusions (NFκB2-ROS1, NFκB2-TYK2, NCOC2-ROS1, and PABPC4-TYK2), some of which appear to activate the JAK/STAT3 pathway (NFκB2-ROS1 and NFκB2-TYK2).
Patients with these fusions did not have JAK1 or STAT3 mutations, which suggests the fusions are an independent cause of ALK-negative ALCL.
To confirm whether JAK1 and STAT3 mutations can cause ALK-negative ALCL, the investigators induced these mutations in normal human cells. The mutations did, in fact, lead to diseased cells.
Finally, the team tested JAK/STAT3 pathway inhibitors—ruxolitinib and PUH71—in mouse models of ALK-negative ALCL. And they found that both drugs significantly inhibited tumor growth.
“Our findings demonstrate that drugs targeting the JAK/STAT3 pathway offer a viable therapeutic strategy in a subset of patients with ALCL,” Dr Rabadan said.
“A couple of JAK/STAT3 inhibitors have been approved by the FDA for the treatment of psoriasis and rheumatoid arthritis, and several more are currently in clinical trials. These could be tested in patients whose genetic profile matches those we identified in our study.” ![]()
Combo improves PFS in untreated CLL
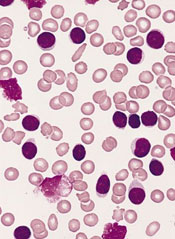
Results of a phase 3 study suggest that adding ofatumumab to chlorambucil can improve progression-free survival (PFS) in treatment-naïve patients with chronic lymphocytic leukemia (CLL).
Ofatumumab plus chlorambucil improved the median PFS by 71% compared to chlorambucil alone.
The combination also improved the overall response rate, duration of response, and time to next treatment.
However, patients in the combination arm had a higher rate of grade 3 or greater adverse events (AEs).
Researchers reported these results in The Lancet. The study, known as COMPLEMENT 1, was funded by GlaxoSmithKline and Genmab A/S.
The study included 447 patients with previously untreated CLL for whom fludarabine-based therapy was considered inappropriate. Patients were randomized to treatment with up to 12 cycles of ofatumumab in combination with chlorambucil (n=221) or up to 12 cycles of chlorambucil alone (n=226).
The study’s primary endpoint was the median PFS, which was 22.4 months in the combination arm and 13.1 months in the chlorambucil arm (hazard ratio [HR]=0.57, P<0.0001). This improvement in PFS was observed in most subgroups, irrespective of age, gender, disease stage, and prognostic factors.
As for secondary endpoints, patients in the combination arm had a higher overall response rate than patients in the chlorambucil arm—82% and 69%, respectively (odds ratio=2.16, P=0.001).
And combination treatment increased the duration of response compared to chlorambucil alone—22.1 months and 13.2 months, respectively (HR=0.56, P<0.001).
Patients in the combination arm also experienced a significantly longer time to next therapy compared to the chlorambucil arm—39.8 months and 24.7 months, respectively (HR=0.49, P<0.0001).
Safety data
The most common AEs (occurring in at least 2% of patients) were neutropenia, thrombocytopenia, anemia, infections, and infusion-related reactions.
Neutropenia occurred more frequently in the combination arm (27% vs 18%), as did infusion-related reactions (67% vs 0%) and infections (46% vs 42%). But thrombocytopenia and anemia were more frequent in the chlorambucil arm (26% vs 14% and 13% vs 9%, respectively).
The incidence of grade 3 or greater AEs was higher in the combination arm than the chlorambucil arm—50% and 43%, respectively.
Grade 3/4 infusion-related reactions occurred in 10% of patients in the combination arm, leading to drug withdrawal in 3% of patients and hospitalization in 2% of patients. No fatal infusion-related reactions were reported.
The most common infections were respiratory tract infections, with an incidence of 27% in the combination arm and 31% in the chlorambucil arm. There were similar frequencies of sepsis (3% and 2%, respectively) and opportunistic infections between the arms (4% and 5%, respectively).
The incidence of AEs leading to treatment withdrawal was 13% in both arms. And the incidence of death during treatment or within 60 days after the last dose was 3% in both arms.
These data formed the basis for regulatory approvals of ofatumumab (Arzerra) in the US and European Union, as well as the recent inclusion of ofatumumab plus chlorambucil in the National Comprehensive Cancer Network treatment guidelines. ![]()

Results of a phase 3 study suggest that adding ofatumumab to chlorambucil can improve progression-free survival (PFS) in treatment-naïve patients with chronic lymphocytic leukemia (CLL).
Ofatumumab plus chlorambucil improved the median PFS by 71% compared to chlorambucil alone.
The combination also improved the overall response rate, duration of response, and time to next treatment.
However, patients in the combination arm had a higher rate of grade 3 or greater adverse events (AEs).
Researchers reported these results in The Lancet. The study, known as COMPLEMENT 1, was funded by GlaxoSmithKline and Genmab A/S.
The study included 447 patients with previously untreated CLL for whom fludarabine-based therapy was considered inappropriate. Patients were randomized to treatment with up to 12 cycles of ofatumumab in combination with chlorambucil (n=221) or up to 12 cycles of chlorambucil alone (n=226).
The study’s primary endpoint was the median PFS, which was 22.4 months in the combination arm and 13.1 months in the chlorambucil arm (hazard ratio [HR]=0.57, P<0.0001). This improvement in PFS was observed in most subgroups, irrespective of age, gender, disease stage, and prognostic factors.
As for secondary endpoints, patients in the combination arm had a higher overall response rate than patients in the chlorambucil arm—82% and 69%, respectively (odds ratio=2.16, P=0.001).
And combination treatment increased the duration of response compared to chlorambucil alone—22.1 months and 13.2 months, respectively (HR=0.56, P<0.001).
Patients in the combination arm also experienced a significantly longer time to next therapy compared to the chlorambucil arm—39.8 months and 24.7 months, respectively (HR=0.49, P<0.0001).
Safety data
The most common AEs (occurring in at least 2% of patients) were neutropenia, thrombocytopenia, anemia, infections, and infusion-related reactions.
Neutropenia occurred more frequently in the combination arm (27% vs 18%), as did infusion-related reactions (67% vs 0%) and infections (46% vs 42%). But thrombocytopenia and anemia were more frequent in the chlorambucil arm (26% vs 14% and 13% vs 9%, respectively).
The incidence of grade 3 or greater AEs was higher in the combination arm than the chlorambucil arm—50% and 43%, respectively.
Grade 3/4 infusion-related reactions occurred in 10% of patients in the combination arm, leading to drug withdrawal in 3% of patients and hospitalization in 2% of patients. No fatal infusion-related reactions were reported.
The most common infections were respiratory tract infections, with an incidence of 27% in the combination arm and 31% in the chlorambucil arm. There were similar frequencies of sepsis (3% and 2%, respectively) and opportunistic infections between the arms (4% and 5%, respectively).
The incidence of AEs leading to treatment withdrawal was 13% in both arms. And the incidence of death during treatment or within 60 days after the last dose was 3% in both arms.
These data formed the basis for regulatory approvals of ofatumumab (Arzerra) in the US and European Union, as well as the recent inclusion of ofatumumab plus chlorambucil in the National Comprehensive Cancer Network treatment guidelines. ![]()

Results of a phase 3 study suggest that adding ofatumumab to chlorambucil can improve progression-free survival (PFS) in treatment-naïve patients with chronic lymphocytic leukemia (CLL).
Ofatumumab plus chlorambucil improved the median PFS by 71% compared to chlorambucil alone.
The combination also improved the overall response rate, duration of response, and time to next treatment.
However, patients in the combination arm had a higher rate of grade 3 or greater adverse events (AEs).
Researchers reported these results in The Lancet. The study, known as COMPLEMENT 1, was funded by GlaxoSmithKline and Genmab A/S.
The study included 447 patients with previously untreated CLL for whom fludarabine-based therapy was considered inappropriate. Patients were randomized to treatment with up to 12 cycles of ofatumumab in combination with chlorambucil (n=221) or up to 12 cycles of chlorambucil alone (n=226).
The study’s primary endpoint was the median PFS, which was 22.4 months in the combination arm and 13.1 months in the chlorambucil arm (hazard ratio [HR]=0.57, P<0.0001). This improvement in PFS was observed in most subgroups, irrespective of age, gender, disease stage, and prognostic factors.
As for secondary endpoints, patients in the combination arm had a higher overall response rate than patients in the chlorambucil arm—82% and 69%, respectively (odds ratio=2.16, P=0.001).
And combination treatment increased the duration of response compared to chlorambucil alone—22.1 months and 13.2 months, respectively (HR=0.56, P<0.001).
Patients in the combination arm also experienced a significantly longer time to next therapy compared to the chlorambucil arm—39.8 months and 24.7 months, respectively (HR=0.49, P<0.0001).
Safety data
The most common AEs (occurring in at least 2% of patients) were neutropenia, thrombocytopenia, anemia, infections, and infusion-related reactions.
Neutropenia occurred more frequently in the combination arm (27% vs 18%), as did infusion-related reactions (67% vs 0%) and infections (46% vs 42%). But thrombocytopenia and anemia were more frequent in the chlorambucil arm (26% vs 14% and 13% vs 9%, respectively).
The incidence of grade 3 or greater AEs was higher in the combination arm than the chlorambucil arm—50% and 43%, respectively.
Grade 3/4 infusion-related reactions occurred in 10% of patients in the combination arm, leading to drug withdrawal in 3% of patients and hospitalization in 2% of patients. No fatal infusion-related reactions were reported.
The most common infections were respiratory tract infections, with an incidence of 27% in the combination arm and 31% in the chlorambucil arm. There were similar frequencies of sepsis (3% and 2%, respectively) and opportunistic infections between the arms (4% and 5%, respectively).
The incidence of AEs leading to treatment withdrawal was 13% in both arms. And the incidence of death during treatment or within 60 days after the last dose was 3% in both arms.
These data formed the basis for regulatory approvals of ofatumumab (Arzerra) in the US and European Union, as well as the recent inclusion of ofatumumab plus chlorambucil in the National Comprehensive Cancer Network treatment guidelines.
System can diagnose lymphoma, other diseases

Photo by Daniel Sone
Scientists say a smartphone-based system could bring rapid, accurate molecular diagnosis of cancers and other diseases to locations lacking the latest medical technology.
In PNAS, the group explained how the digital diffraction diagnosis (D3) system collects detailed microscopic images for digital analysis of the molecular composition of cells and tissues.
In pilot experiments, the system enabled accurate diagnoses of lymphoma and cervical cancer.
“The emerging genomic and biological data for various cancers, which can be essential to choosing the most appropriate therapy, supports the need for molecular profiling strategies that are more accessible to providers, clinical investigators, and patients,” said study author Cesar Castro, MD, of Massachusetts General Hospital in Boston.
“And we believe the platform we have developed provides essential features at an extraordinarily low cost.”
The D3 system features an imaging module with a battery-powered LED light clipped onto a standard smartphone that records high-resolution imaging data with its camera.
With a greater field of view than traditional microscopy, the D3 system is capable of recording data on more than 100,000 cells from a blood or tissue sample in a single image. The data can then be transmitted for analysis to a remote graphic-processing server via a secure, encrypted cloud service, and the results returned to the point of care.
For molecular analysis of tumors, a sample of blood or tissue is labeled with microbeads that bind to known cancer-related molecules and loaded into the D3 imaging module.
After the image is recorded and data transmitted to the server, the presence of specific molecules is detected by analyzing the diffraction patterns generated by the microbeads. The use of variously sized or coated beads may offer unique diffraction signatures to facilitate detection.
A numerical algorithm the researchers developed can distinguish cells from beads and analyze as much as 10 MB of data in less than nine hundredths of a second.
In a pilot test with cancer cell lines, the D3 system detected the presence of tumor proteins with an accuracy matching that of the current gold standard for molecular profiling. And the system’s larger field of view enabled simultaneous analysis of more than 100,000 cells at a time.
The researchers also conducted analyses of cervical biopsy samples from 25 women with abnormal PAP smears—samples collected along with those used for clinical diagnosis—using microbeads tagged with antibodies against 3 published markers of cervical cancer.
Based on the number of antibody-tagged microbeads binding to cells, D3 analysis promptly and reliably categorized biopsy samples as high-risk, low-risk, or benign. Results matched those of conventional pathologic analysis.
In addition, D3 analysis of fine-needle lymph node biopsy samples was accurately able to differentiate 4 patients whose lymphoma diagnosis was confirmed by conventional pathology from another 4 patients with benign lymph node enlargement.
Along with protein analyses, the D3 system was enhanced to successfully detect DNA—in this instance, from human papilloma virus—with great sensitivity.
In all of these tests, results were available in under an hour and at a cost of $1.80 per assay, a price that would be expected to drop with further refinement of the D3 system.
“We expect that the D3 platform will enhance the breadth and depth of cancer screening in a way that is feasible and sustainable for resource limited-settings,” said Ralph Weissleder, MD, PhD, also of Massachusetts General Hospital.
“By taking advantage of the increased penetration of mobile phone technology worldwide, the system should allow the prompt triaging of suspicious or high-risk cases that could help to offset delays caused by limited pathology services in those regions and reduce the need for patients to return for follow-up care, which is often challenging for them.”
The researchers’ next steps are to investigate D3’s ability to analyze protein and DNA markers of other disease catalysts, integrate the software with larger databases, and conduct clinical studies in settings such as care-delivery sites in developing countries or rural areas.
Massachusetts General Hospital has filed a patent application covering the D3 technology.

Photo by Daniel Sone
Scientists say a smartphone-based system could bring rapid, accurate molecular diagnosis of cancers and other diseases to locations lacking the latest medical technology.
In PNAS, the group explained how the digital diffraction diagnosis (D3) system collects detailed microscopic images for digital analysis of the molecular composition of cells and tissues.
In pilot experiments, the system enabled accurate diagnoses of lymphoma and cervical cancer.
“The emerging genomic and biological data for various cancers, which can be essential to choosing the most appropriate therapy, supports the need for molecular profiling strategies that are more accessible to providers, clinical investigators, and patients,” said study author Cesar Castro, MD, of Massachusetts General Hospital in Boston.
“And we believe the platform we have developed provides essential features at an extraordinarily low cost.”
The D3 system features an imaging module with a battery-powered LED light clipped onto a standard smartphone that records high-resolution imaging data with its camera.
With a greater field of view than traditional microscopy, the D3 system is capable of recording data on more than 100,000 cells from a blood or tissue sample in a single image. The data can then be transmitted for analysis to a remote graphic-processing server via a secure, encrypted cloud service, and the results returned to the point of care.
For molecular analysis of tumors, a sample of blood or tissue is labeled with microbeads that bind to known cancer-related molecules and loaded into the D3 imaging module.
After the image is recorded and data transmitted to the server, the presence of specific molecules is detected by analyzing the diffraction patterns generated by the microbeads. The use of variously sized or coated beads may offer unique diffraction signatures to facilitate detection.
A numerical algorithm the researchers developed can distinguish cells from beads and analyze as much as 10 MB of data in less than nine hundredths of a second.
In a pilot test with cancer cell lines, the D3 system detected the presence of tumor proteins with an accuracy matching that of the current gold standard for molecular profiling. And the system’s larger field of view enabled simultaneous analysis of more than 100,000 cells at a time.
The researchers also conducted analyses of cervical biopsy samples from 25 women with abnormal PAP smears—samples collected along with those used for clinical diagnosis—using microbeads tagged with antibodies against 3 published markers of cervical cancer.
Based on the number of antibody-tagged microbeads binding to cells, D3 analysis promptly and reliably categorized biopsy samples as high-risk, low-risk, or benign. Results matched those of conventional pathologic analysis.
In addition, D3 analysis of fine-needle lymph node biopsy samples was accurately able to differentiate 4 patients whose lymphoma diagnosis was confirmed by conventional pathology from another 4 patients with benign lymph node enlargement.
Along with protein analyses, the D3 system was enhanced to successfully detect DNA—in this instance, from human papilloma virus—with great sensitivity.
In all of these tests, results were available in under an hour and at a cost of $1.80 per assay, a price that would be expected to drop with further refinement of the D3 system.
“We expect that the D3 platform will enhance the breadth and depth of cancer screening in a way that is feasible and sustainable for resource limited-settings,” said Ralph Weissleder, MD, PhD, also of Massachusetts General Hospital.
“By taking advantage of the increased penetration of mobile phone technology worldwide, the system should allow the prompt triaging of suspicious or high-risk cases that could help to offset delays caused by limited pathology services in those regions and reduce the need for patients to return for follow-up care, which is often challenging for them.”
The researchers’ next steps are to investigate D3’s ability to analyze protein and DNA markers of other disease catalysts, integrate the software with larger databases, and conduct clinical studies in settings such as care-delivery sites in developing countries or rural areas.
Massachusetts General Hospital has filed a patent application covering the D3 technology.

Photo by Daniel Sone
Scientists say a smartphone-based system could bring rapid, accurate molecular diagnosis of cancers and other diseases to locations lacking the latest medical technology.
In PNAS, the group explained how the digital diffraction diagnosis (D3) system collects detailed microscopic images for digital analysis of the molecular composition of cells and tissues.
In pilot experiments, the system enabled accurate diagnoses of lymphoma and cervical cancer.
“The emerging genomic and biological data for various cancers, which can be essential to choosing the most appropriate therapy, supports the need for molecular profiling strategies that are more accessible to providers, clinical investigators, and patients,” said study author Cesar Castro, MD, of Massachusetts General Hospital in Boston.
“And we believe the platform we have developed provides essential features at an extraordinarily low cost.”
The D3 system features an imaging module with a battery-powered LED light clipped onto a standard smartphone that records high-resolution imaging data with its camera.
With a greater field of view than traditional microscopy, the D3 system is capable of recording data on more than 100,000 cells from a blood or tissue sample in a single image. The data can then be transmitted for analysis to a remote graphic-processing server via a secure, encrypted cloud service, and the results returned to the point of care.
For molecular analysis of tumors, a sample of blood or tissue is labeled with microbeads that bind to known cancer-related molecules and loaded into the D3 imaging module.
After the image is recorded and data transmitted to the server, the presence of specific molecules is detected by analyzing the diffraction patterns generated by the microbeads. The use of variously sized or coated beads may offer unique diffraction signatures to facilitate detection.
A numerical algorithm the researchers developed can distinguish cells from beads and analyze as much as 10 MB of data in less than nine hundredths of a second.
In a pilot test with cancer cell lines, the D3 system detected the presence of tumor proteins with an accuracy matching that of the current gold standard for molecular profiling. And the system’s larger field of view enabled simultaneous analysis of more than 100,000 cells at a time.
The researchers also conducted analyses of cervical biopsy samples from 25 women with abnormal PAP smears—samples collected along with those used for clinical diagnosis—using microbeads tagged with antibodies against 3 published markers of cervical cancer.
Based on the number of antibody-tagged microbeads binding to cells, D3 analysis promptly and reliably categorized biopsy samples as high-risk, low-risk, or benign. Results matched those of conventional pathologic analysis.
In addition, D3 analysis of fine-needle lymph node biopsy samples was accurately able to differentiate 4 patients whose lymphoma diagnosis was confirmed by conventional pathology from another 4 patients with benign lymph node enlargement.
Along with protein analyses, the D3 system was enhanced to successfully detect DNA—in this instance, from human papilloma virus—with great sensitivity.
In all of these tests, results were available in under an hour and at a cost of $1.80 per assay, a price that would be expected to drop with further refinement of the D3 system.
“We expect that the D3 platform will enhance the breadth and depth of cancer screening in a way that is feasible and sustainable for resource limited-settings,” said Ralph Weissleder, MD, PhD, also of Massachusetts General Hospital.
“By taking advantage of the increased penetration of mobile phone technology worldwide, the system should allow the prompt triaging of suspicious or high-risk cases that could help to offset delays caused by limited pathology services in those regions and reduce the need for patients to return for follow-up care, which is often challenging for them.”
The researchers’ next steps are to investigate D3’s ability to analyze protein and DNA markers of other disease catalysts, integrate the software with larger databases, and conduct clinical studies in settings such as care-delivery sites in developing countries or rural areas.
Massachusetts General Hospital has filed a patent application covering the D3 technology.